| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:18:58 UTC |
|---|
| Updated at | 2020-11-18 16:39:30 UTC |
|---|
| CannabisDB ID | CDB005196 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Pyridoxal 5'-phosphate |
|---|
| Description | Pyridoxal 5'-phosphate, also known as PLP or codecarboxylase, belongs to the class of organic compounds known as pyridoxals and derivatives. Pyridoxals and derivatives are compounds containing a pyridoxal moiety, which consists of a pyridine ring substituted at positions 2,3,4, and 5 by a methyl group, a hydroxyl group, a carbaldehyde group, and a hydroxymethyl group, respectively. Pyridoxal 5'-phosphate is a drug which is used for nutritional supplementation and for treating dietary shortage or imbalance. Pyridoxal 5'-phosphate is a strong basic compound (based on its pKa). Pyridoxal 5'-phosphate exists in all living species, ranging from bacteria to humans. In humans, pyridoxal 5'-phosphate is involved in glycine and serine metabolism. Outside of the human body, Pyridoxal 5'-phosphate is found, on average, in the highest concentration within milk (cow). Pyridoxal 5'-phosphate has also been detected, but not quantified in, several different foods, such as soursops, italian sweet red peppers, muscadine grapes, european plums, and blackcurrants. This could make pyridoxal 5'-phosphate a potential biomarker for the consumption of these foods. The monophosphate ester obtained by condensation of phosphoric acid with the primary hydroxy group of pyridoxal. Pyridoxal 5'-phosphate is a potentially toxic compound. Pyridoxal 5'-phosphate, with regard to humans, has been found to be associated with several diseases such as epilepsy, early-onset, vitamin b6-dependent, odontohypophosphatasia, pyridoxamine 5-prime-phosphate oxidase deficiency, and hypophosphatasia; pyridoxal 5'-phosphate has also been linked to the inborn metabolic disorder celiac disease. Pyridoxal 5'-phosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Hydroxy-2-methyl-5-[(phosphonooxy)methyl]-4-pyridinecarboxaldehyde | ChEBI | | 3-Hydroxy-5-(hydroxymethyl)-2-methylisonicotinaldehyde 5-phosphate | ChEBI | | Codecarboxylase | ChEBI | | Phosphoric acid mono-(4-formyl-5-hydroxy-6-methyl-pyridin-3-ylmethyl) ester | ChEBI | | PLP | ChEBI | | Pyridoxal 5'-(dihydrogen phosphate) | ChEBI | | Pyridoxal 5-monophosphoric acid ester | ChEBI | | Pyridoxal 5-phosphate | ChEBI | | Pyridoxal phosphate | ChEBI | | PYRIDOXAL-5'-phosphATE | ChEBI | | 3-Hydroxy-5-(hydroxymethyl)-2-methylisonicotinaldehyde 5-phosphoric acid | Generator | | Phosphate mono-(4-formyl-5-hydroxy-6-methyl-pyridin-3-ylmethyl) ester | Generator | | Pyridoxal 5'-(dihydrogen phosphoric acid) | Generator | | Pyridoxal 5-monophosphate ester | Generator | | Pyridoxal 5-phosphoric acid | Generator | | Pyridoxal phosphoric acid | Generator | | PYRIDOXAL-5'-phosphoric acid | Generator | | Pyridoxal 5'-phosphoric acid | Generator | | Apolon b6 | HMDB | | Biosechs | HMDB | | Coenzyme b6 | HMDB | | Hairoxal | HMDB | | Hexermin-p | HMDB | | Hi-pyridoxin | HMDB | | Hiadelon | HMDB | | Himitan | HMDB | | PAL-p | HMDB | | Phosphopyridoxal | HMDB | | Phosphopyridoxal coenzyme | HMDB | | Pidopidon | HMDB | | Piodel | HMDB | | Pydoxal | HMDB | | Pyridoxal p | HMDB | | Pyridoxal-p | HMDB | | Pyridoxyl phosphate | HMDB | | Pyromijin | HMDB | | Sechvitan | HMDB | | Vitahexin-p | HMDB | | Vitazechs | HMDB | | Phosphate, pyridoxal | HMDB | | Pyridoxal 5 phosphate | HMDB |
|
|---|
| Chemical Formula | C8H10NO6P |
|---|
| Average Molecular Weight | 247.14 |
|---|
| Monoisotopic Molecular Weight | 247.0246 |
|---|
| IUPAC Name | [(4-formyl-5-hydroxy-6-methylpyridin-3-yl)methoxy]phosphonic acid |
|---|
| Traditional Name | pyridoxal phosphate |
|---|
| CAS Registry Number | 54-47-7 |
|---|
| SMILES | CC1=NC=C(COP(O)(O)=O)C(C=O)=C1O |
|---|
| InChI Identifier | InChI=1S/C8H10NO6P/c1-5-8(11)7(3-10)6(2-9-5)4-15-16(12,13)14/h2-3,11H,4H2,1H3,(H2,12,13,14) |
|---|
| InChI Key | NGVDGCNFYWLIFO-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pyridoxals and derivatives. Pyridoxals and derivatives are compounds containing a pyridoxal moiety, which consists of a pyridine ring substituted at positions 2,3,4, and 5 by a methyl group, a hydroxyl group, a carbaldehyde group, and a hydroxymethyl group, respectively. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pyridines and derivatives |
|---|
| Sub Class | Pyridine carboxaldehydes |
|---|
| Direct Parent | Pyridoxals and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyridoxal
- Aryl-aldehyde
- Monoalkyl phosphate
- Hydroxypyridine
- Methylpyridine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Vinylogous acid
- Heteroaromatic compound
- Azacycle
- Organopnictogen compound
- Aldehyde
- Organic oxygen compound
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 255 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 28 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Pyridoxal 5'-phosphate, 1 MEOX; 3 TMS, GC-MS Spectrum | splash10-0gb9-2690000000-c9bacb7e657461e28407 | Spectrum | | GC-MS | Pyridoxal 5'-phosphate, non-derivatized, GC-MS Spectrum | splash10-0gb9-2690000000-c9bacb7e657461e28407 | Spectrum | | GC-MS | Pyridoxal 5'-phosphate, non-derivatized, GC-MS Spectrum | splash10-004j-0910000000-0408f3957221fc9882cd | Spectrum | | GC-MS | Pyridoxal 5'-phosphate, non-derivatized, GC-MS Spectrum | splash10-0gb9-1790000000-b01115fa8b4cfd3612c7 | Spectrum | | Predicted GC-MS | Pyridoxal 5'-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-9520000000-bfd574b2e5355694902a | Spectrum | | Predicted GC-MS | Pyridoxal 5'-phosphate, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-006t-9152000000-26c9f631195663322986 | Spectrum | | Predicted GC-MS | Pyridoxal 5'-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0f6t-0690000000-4aa57f3f28f0bdef9dcf | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0fxx-8900000000-ee972556340c61650528 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-014l-9100000000-3cc29fb51820adecd59c | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0090000000-9b86e80a9dd0de14ab59 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-03dj-9800000000-9313aa9cbf19bc6311c5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0002-9500000000-7ce120a58879e2214ce5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-004j-9200000000-7e6bd8c298613e59fb5d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-004i-9000000000-4c05178b1645bf01f3fa | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-3920000000-6c5e6106f5658d1d6c50 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-3920000000-f96097815e6922356131 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-002b-9000000000-2c4e243699a95e3a0f88 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-002b-9000000000-8aa111485891af9f0d02 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-0090000000-9b86e80a9dd0de14ab59 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03dj-9800000000-9313aa9cbf19bc6311c5 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-9500000000-7ce120a58879e2214ce5 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004j-9200000000-7e6bd8c298613e59fb5d | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-4c05178b1645bf01f3fa | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-002b-9000000000-2c4e243699a95e3a0f88 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-002b-9000000000-8aa111485891af9f0d02 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f6t-1790000000-1dcb07cd1110c71f7005 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-ca6aab7f31a36863a417 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9800000000-053802b9c0533d7943ea | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9080000000-12ca63d34d39d59f6b42 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-144099ec201adbbc4684 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-70a9559d65e78c488e7e | 2015-05-27 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Glucose-Alanine Cycle | 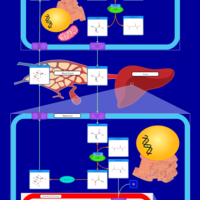 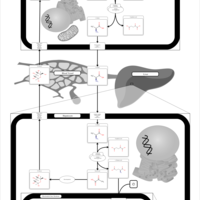 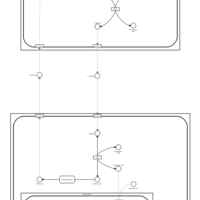 | Not Available | | Malate-Aspartate Shuttle |  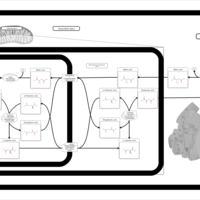 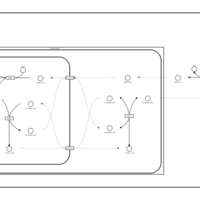 | Not Available | | Vitamin B6 Metabolism | 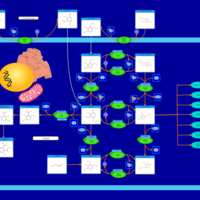 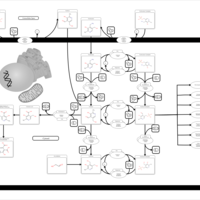 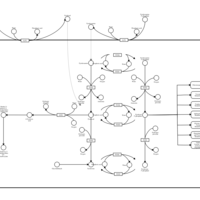 |  | | Alanine Metabolism | 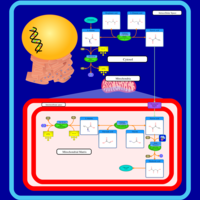 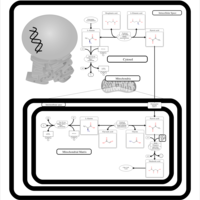  |  | | Primary Hyperoxaluria Type I | 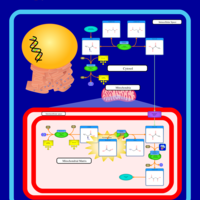 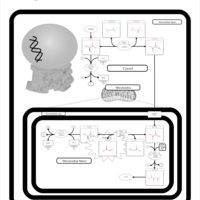  | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| 5-aminolevulinate synthase, erythroid-specific, mitochondrial | ALAS2 | Xp11.21 | P22557 | details | | 5-aminolevulinate synthase, nonspecific, mitochondrial | ALAS1 | 3p21.1 | P13196 | details | | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial | GCAT | 22q13.1 | O75600 | details | | Methylenetetrahydrofolate reductase | MTHFR | 1p36.3 | P42898 | details | | 4-aminobutyrate aminotransferase, mitochondrial | ABAT | 16p13.2 | P80404 | details | | Tyrosine aminotransferase | TAT | 16q22.1 | P17735 | details | | Aromatic-L-amino-acid decarboxylase | DDC | 7p12.2 | P20711 | details | | Aspartate aminotransferase, cytoplasmic | GOT1 | 10q24.1-q25.1 | P17174 | details | | Aspartate aminotransferase, mitochondrial | GOT2 | 16q21 | P00505 | details | | Branched-chain-amino-acid aminotransferase, cytosolic | BCAT1 | 12p12.1 | P54687 | details | | Kynureninase | KYNU | 2q22.2 | Q16719 | details | | Glucose-6-phosphate 1-dehydrogenase | G6PD | Xq28 | P11413 | details | | Glycogen phosphorylase, liver form | PYGL | 14q21-q22 | P06737 | details | | Glycogen phosphorylase, muscle form | PYGM | 11q12-q13.2 | P11217 | details | | Glycogen phosphorylase, brain form | PYGB | 20p11.21 | P11216 | details | | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | AADAT | 4q33 | Q8N5Z0 | details | | Alanine--glyoxylate aminotransferase 2, mitochondrial | AGXT2 | 5p13 | Q9BYV1 | details | | Ornithine aminotransferase, mitochondrial | OAT | 10q26 | P04181 | details | | Kynurenine--oxoglutarate transaminase 1 | CCBL1 | 9q34.11 | Q16773 | details | | Branched-chain-amino-acid aminotransferase, mitochondrial | BCAT2 | 19q13 | O15382 | details | | Cystathionine gamma-lyase | CTH | 1p31.1 | P32929 | details | | L-serine dehydratase/L-threonine deaminase | SDS | 12q24.13 | P20132 | details | | Pyridoxine-5'-phosphate oxidase | PNPO | 17q21.32 | Q9NVS9 | details | | Formimidoyltransferase-cyclodeaminase | FTCD | 21q22.3 | O95954 | details | | Glutamate decarboxylase 2 | GAD2 | 10p11.23 | Q05329 | details | | Glutamate decarboxylase 1 | GAD1 | 2q31 | Q99259 | details | | Cystathionine beta-synthase | CBS | 21q22.3 | P35520 | details | | Serine--pyruvate aminotransferase | AGXT | 2q37.3 | P21549 | details | | Serine hydroxymethyltransferase, mitochondrial | SHMT2 | 12q12-q14 | P34897 | details | | Serine hydroxymethyltransferase, cytosolic | SHMT1 | 17p11.2 | P34896 | details | | Glycine dehydrogenase [decarboxylating], mitochondrial | GLDC | 9p22 | P23378 | details | | Alanine aminotransferase 1 | GPT | 8q24.3 | P24298 | details | | Phosphoserine aminotransferase | PSAT1 | 9q21.2 | Q9Y617 | details | | Histidine decarboxylase | HDC | 15q21-q22 | P19113 | details | | Serine palmitoyltransferase 1 | SPTLC1 | 9q22.2 | O15269 | details | | Serine palmitoyltransferase 2 | SPTLC2 | 14q24.3 | O15270 | details | | Cysteine sulfinic acid decarboxylase | CSAD | 12q13.11-q14.3 | Q9Y600 | details | | Ornithine decarboxylase | ODC1 | 2p25 | P11926 | details | | Sphingosine-1-phosphate lyase 1 | SGPL1 | 10q21 | O95470 | details | | Pyridoxal kinase | PDXK | 21q22.3 | O00764 | details | | Arginine decarboxylase | ADC | 1p35.1 | Q96A70 | details | | Phosphatidylserine decarboxylase proenzyme | PISD | 22q12.2 | Q9UG56 | details | | Cysteine desulfurase, mitochondrial | NFS1 | 20q11.22 | Q9Y697 | details | | Proline synthase co-transcribed bacterial homolog protein | PROSC | 8p11.2 | O94903 | details | | Aminoadipate aminotransferase, isoform CRA_b | AADAT | 4q33 | Q4W5N8 | details | | Threonine synthase-like 1 | THNSL1 | 10p12.1 | Q8IYQ7 | details | | Ethanolamine-phosphate phospho-lyase | AGXT2L1 | 4q25 | Q8TBG4 | details | | Alanine aminotransferase 2 | GPT2 | 16q12.1 | Q8TD30 | details | | Pyridoxal phosphate phosphatase | PDXP | 22q12.3 | Q96GD0 | details | | Selenocysteine lyase | SCLY | 2q37.3 | Q96I15 | details | | Pyridoxal phosphate phosphatase PHOSPHO2 | PHOSPHO2 | 2q31.1 | Q8TCD6 | details | | Putative aspartate aminotransferase, cytoplasmic 2 | GOT1L1 | 8p11.23 | Q8NHS2 | details | | 5-phosphohydroxy-L-lysine phospho-lyase | AGXT2L2 | 5q35.3 | Q8IUZ5 | details | | Glutamate decarboxylase-like protein 1 | GADL1 | 3p24.1-p23 | Q6ZQY3 | details | | Kynurenine--oxoglutarate transaminase 3 | CCBL2 | 1p22.2 | Q6YP21 | details | | Molybdenum cofactor sulfurase | MOCOS | 18q12 | Q96EN8 | details | | Pyridoxal-dependent decarboxylase domain-containing protein 1 | PDXDC1 | 16p13.11 | Q6P996 | details | | Pyridoxal-dependent decarboxylase domain-containing protein 2 | PDXDC2 | 16q22.1 | Q6P474 | details | | Serine dehydratase-like | SDSL | 12q24.13 | Q96GA7 | details | | O-phosphoseryl-tRNA(Sec) selenium transferase | SEPSECS | 4p15.2 | Q9HD40 | details | | Serine palmitoyltransferase 3 | SPTLC3 | 20p12.1 | Q9NUV7 | details | | Serine racemase | SRR | 17p13 | Q9GZT4 | details | | Threonine synthase-like 2 | THNSL2 | 2p11.2 | Q86YJ6 | details | | Serine hydroxymethyltransferase | SHMT1 | 17p11.2 | A8MYA6 | details | | Aspartate aminotransferase | GIG18 | | Q2TU84 | details | | GAD1 protein | GAD1 | 2q31 | Q49AK1 | details | | Dopa decarboxylase (Aromatic L-amino acid decarboxylase) | DDC | 7p12.2 | Q53Y41 | details | | Phosphorylase | | | Q59GM9 | details | | Aminolevulinate, delta-, synthase 1, isoform CRA_a | ALAS1 | 3p21.1 | Q5JAM2 | details | | Aminolevulinate, delta-, synthase 2 | ALAS2 | | Q5JZF5 | details | | Glutamate decarboxylase 2 (Pancreatic islets and brain, 65kDa) | GAD2 | 10p11.23 | Q5VZ29 | details | | Putative L-Dopa decarboxylase | DDC | 7p12.2 | Q5W5T9 | details | | DDC protein | DDC | 7p12.2 | Q6IBS8 | details | | CSAD protein | CSAD | 12q13.11-q14.3 | Q86V02 | details | | PDXDC1 protein | PDXDC1 | 16p13.11 | Q86XE2 | details | | Glutamate decarboxylase 1 (Brain, 67kDa) | GAD1 | 2q31 | Q8IVA8 | details | | P-selectin cytoplasmic tail-associated protein (PCAP) | pcap | 1q42.2-q43 | Q96JQ3 | details | | Glutamic acid decarboxylase | GAD65 | | Q9UGI5 | details | | Cysteine sulfinic acid decarboxylase, isoform CRA_c | CSAD | 12q13.11-q14.3 | Q9Y602 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001491 |
|---|
| DrugBank ID | DB00114 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB021820 |
|---|
| KNApSAcK ID | C00007503 |
|---|
| Chemspider ID | 1022 |
|---|
| KEGG Compound ID | C00018 |
|---|
| BioCyc ID | PYRIDOXAL_PHOSPHATE |
|---|
| BiGG ID | 33526 |
|---|
| Wikipedia Link | Pyridoxal_phosphate |
|---|
| METLIN ID | 6275 |
|---|
| PubChem Compound | 1051 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 18405 |
|---|
| References |
|---|
| General References | Not Available |
|---|

