| Identification |
|---|
| HMDB Protein ID
| CDBP00796 |
| Secondary Accession Numbers
| Not Available |
| Name
| Phosphoserine aminotransferase |
| Description
| Not Available |
| Synonyms
|
- PSAT
- Phosphohydroxythreonine aminotransferase
|
| Gene Name
| PSAT1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in metabolic process |
| Specific Function
| Catalyzes the reversible conversion of 3-phosphohydroxypyruvate to phosphoserine and of 3-hydroxy-2-oxo-4-phosphonooxybutanoate to phosphohydroxythreonine (By similarity).
|
| GO Classification
|
| Biological Process |
| pyridoxine biosynthetic process |
| L-serine biosynthetic process |
| Cellular Component |
| cytosol |
| Function |
| binding |
| catalytic activity |
| cofactor binding |
| pyridoxal phosphate binding |
| Molecular Function |
| O-phospho-L-serine:2-oxoglutarate aminotransferase activity |
| pyridoxal phosphate binding |
| Process |
| metabolic process |
| cellular metabolic process |
| serine family amino acid metabolic process |
| l-serine metabolic process |
| cellular amino acid and derivative metabolic process |
| cellular amino acid metabolic process |
| l-serine biosynthetic process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | L-serine biosynthesis | Not Available | Not Available | | pyridoxine 5'-phosphate biosynthesis | Not Available | Not Available | | Glycine, serine and threonine metabolism | Not Available |  | | Glycine and Serine Metabolism | 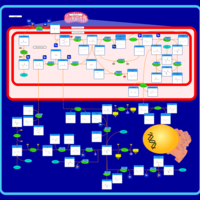 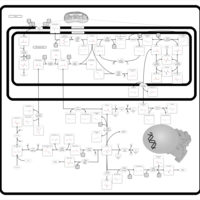 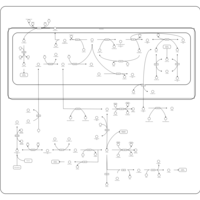 |  | | Dimethylglycine Dehydrogenase Deficiency | 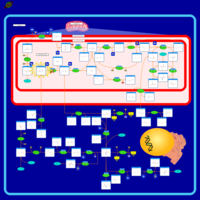 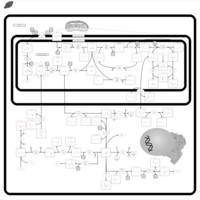  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 9 |
| Locus
| 9q21.2 |
| SNPs
| PSAT1 |
| Gene Sequence
|
>1113 bp
ATGGACGCCCCCAGGCAGGTGGTCAACTTTGGGCCTGGTCCCGCCAAGCTGCCGCACTCA
GTGTTGTTAGAGATACAAAAGGAATTATTAGACTACAAAGGAGTTGGCATTAGTGTTCTT
GAAATGAGTCACAGGTCATCAGATTTTGCCAAGATTATTAACAATACAGAGAATCTTGTG
CGGGAATTGCTAGCTGTTCCAGACAACTATAAGGTGATTTTTCTGCAAGGAGGTGGGTGC
GGCCAGTTCAGTGCTGTCCCCTTAAACCTCATTGGCTTGAAAGCAGGAAGGTGTGCTGAC
TATGTGGTGACAGGAGCTTGGTCAGCTAAGGCCGCAGAAGAAGCCAAGAAGTTTGGGACT
ATAAATATCGTTCACCCTAAACTTGGGAGTTATACAAAAATTCCAGATCCAAGCACCTGG
AACCTCAACCCAGATGCCTCCTACGTGTATTATTGCGCAAATGAGACGGTGCATGGTGTG
GAGTTTGACTTTATACCCGATGTCAAGGGAGCAGTACTGGTTTGTGACATGTCCTCAAAC
TTCCTGTCCAAGCCAGTGGATGTTTCCAAGTTTGGTGTGATTTTTGCTGGTGCCCAGAAG
AATGTTGGCTCTGCTGGGGTCACCGTGGTGATTGTCCGTGATGACCTGCTGGGGTTTGCC
CTCCGAGAGTGCCCCTCGGTCCTGGAATACAAGGTGCAGGCTGGAAACAGCTCCTTGTAC
AACACGCCTCCATGTTTCAGCATCTACGTCATGGGCTTGGTTCTGGAGTGGATTAAAAAC
AATGGAGGTGCCGCGGCCATGGAGAAGCTTAGCTCCATCAAATCTCAAACAATTTATGAG
ATTATTGATAATTCTCAAGGATTCTACGTTTGTCCAGTGGAGCCCCAAAATAGAAGCAAG
ATGAATATTCCATTCCGCATTGGCAATGCCAAAGGAGATGATGCTTTAGAAAAAAGATTT
CTTGATAAAGCTCTTGAACTCAATATGTTGTCCTTGAAAGGGCATAGGTCTGTGGGAGGC
ATCCGGGCCTCTCTGTATAATGCTGTCACAATTGAAGACGTTCAGAAGCTGGCCGCCTTC
ATGAAAAAATTTTTGGAGATGCATCAGCTATGA
|
| Protein Properties |
|---|
| Number of Residues
| 370 |
| Molecular Weight
| 35188.305 |
| Theoretical pI
| 6.68 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Phosphoserine aminotransferase
MDAPRQVVNFGPGPAKLPHSVLLEIQKELLDYKGVGISVLEMSHRSSDFAKIINNTENLV
RELLAVPDNYKVIFLQGGGCGQFSAVPLNLIGLKAGRCADYVVTGAWSAKAAEEAKKFGT
INIVHPKLGSYTKIPDPSTWNLNPDASYVYYCANETVHGVEFDFIPDVKGAVLVCDMSSN
FLSKPVDVSKFGVIFAGAQKNVGSAGVTVVIVRDDLLGFALRECPSVLEYKVQAGNSSLY
NTPPCFSIYVMGLVLEWIKNNGGAAAMEKLSSIKSQTIYEIIDNSQGFYVCPVEPQNRSK
MNIPFRIGNAKGDDALEKRFLDKALELNMLSLKGHRSVGGIRASLYNAVTIEDVQKLAAF
MKKFLEMHQL
|
| External Links |
|---|
| GenBank ID Protein
| 17402893 |
| UniProtKB/Swiss-Prot ID
| Q9Y617 |
| UniProtKB/Swiss-Prot Entry Name
| SERC_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_058179.2 |
| GeneCard ID
| PSAT1 |
| GenAtlas ID
| PSAT1 |
| HGNC ID
| HGNC:19129 |
| References |
|---|
| General References
| Not Available |
