| Identification |
|---|
| HMDB Protein ID
| CDBP02374 |
| Secondary Accession Numbers
| Not Available |
| Name
| Pyridoxal phosphate phosphatase |
| Description
| Not Available |
| Synonyms
|
- PLP phosphatase
- Chronophin
|
| Gene Name
| PDXP |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| Protein serine phosphatase that dephosphorylates 'Ser-3' in cofilin and probably also dephosphorylates phospho-serine residues in DSTN. Regulates cofilin-dependent actin cytoskeleton reorganization. Required for normal progress through mitosis and normal cytokinesis. Does not dephosphorylate phospho-threonines in LIMK1. Does not dephosphorylate peptides containing phospho-tyrosine. Pyridoxal phosphate phosphatase. Has some activity towards pyridoxal 5'-phosphate (PLP), pyridoxine 5'-phosphate (PMP) and pyridoxine 5'-phosphate (PNP), with a highest activity with PLP followed by PNP.
|
| GO Classification
|
| Biological Process |
| actin rod assembly |
| cellular response to ATP |
| positive regulation of actin filament depolymerization |
| regulation of cytokinesis |
| regulation of mitosis |
| protein dephosphorylation |
| Cellular Component |
| cytosol |
| cytoskeleton |
| ruffle membrane |
| lamellipodium membrane |
| Function |
| hydrolase activity, acting on ester bonds |
| catalytic activity |
| hydrolase activity |
| phosphoric ester hydrolase activity |
| phosphatase activity |
| Molecular Function |
| metal ion binding |
| pyridoxal phosphatase activity |
| heat shock protein binding |
| phosphoserine phosphatase activity |
| phosphoprotein phosphatase activity |
| Process |
| metabolic process |
|
| Cellular Location
|
- Cytoplasmic
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Vitamin B6 Metabolism | 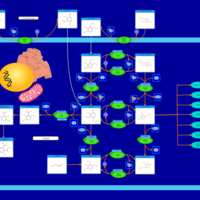 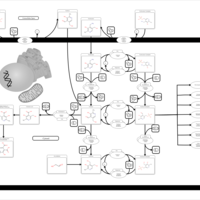 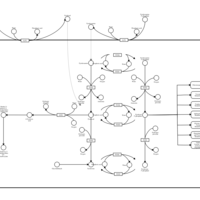 |  | | Hypophosphatasia | 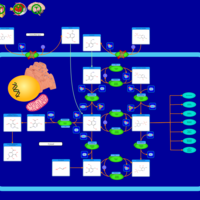 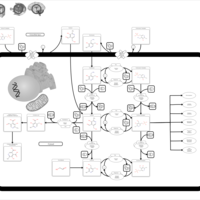 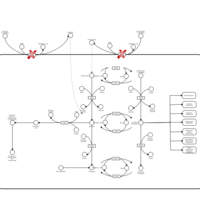 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 22 |
| Locus
| 22q12.3 |
| SNPs
| PDXP |
| Gene Sequence
|
>891 bp
ATGGCGCGCTGCGAGAGGCTGCGCGGAGCGGCCCTGCGCGACGTGCTGGGCCGGGCGCAG
GGGGTCCTGTTCGACTGTGACGGGGTGCTGTGGAACGGCGAGCGCGCCGTGCCGGGCGCC
CCGGAGCTGCTGGAGCGGCTGGCGCGGGCCGGCAAGGCGGCTCTGTTTGTGAGCAACAAC
AGCCGGCGCGCGCGGCCCGAGCTGGCCCTGCGCTTCGCGCGCCTCGGCTTCGGGGGGCTG
CGCGCCGAGCAGCTCTTCAGCTCCGCGCTGTGCGCCGCGCGCCTGCTGCGCCAGCGCCTG
CCCGGGCCTCCGGACGCGCCGGGCGCCGTGTTCGTGCTGGGCGGCGAGGGGCTGCGCGCC
GAGCTGCGCGCCGCGGGGCTGCGCCTGGCCGGGGACCCGAGCGCGGGGGACGGCGCGGCC
CCGCGCGTGCGCGCCGTGCTTGTGGGCTACGACGAGCACTTCTCCTTCGCCAAGCTGAGG
GAGGCGTGCGCGCACCTGCGCGACCCCGAGTGCCTACTCGTGGCCACCGACCGTGACCCA
TGGCACCCGCTGAGCGACGGCAGCCGGACCCCTGGCACCGGGAGCCTGGCCGCTGCAGTG
GAGACAGCCTCGGGACGCCAGGCCCTGGTGGTGGGCAAGCCCAGCCCCTACATGTTCGAG
TGCATCACGGAGAACTTCAGCATCGACCCCGCACGCACGCTTATGGTGGGTGACCGCCTG
GAGACCGACATCCTCTTTGGCCACCGCTGCGGCATGACCACTGTGCTCACGCTCACAGGA
GTCTCCCGCCTAGAAGAGGCCCAGGCCTACCTAGCGGCCGGCCAGCACGACCTCGTGCCC
CATTACTATGTGGAGAGCATCGCAGACTTGACAGAGGGGTTGGAGGACTGA
|
| Protein Properties |
|---|
| Number of Residues
| 296 |
| Molecular Weight
| 31697.735 |
| Theoretical pI
| 6.544 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Pyridoxal phosphate phosphatase
MARCERLRGAALRDVLGRAQGVLFDCDGVLWNGERAVPGAPELLERLARAGKAALFVSNN
SRRARPELALRFARLGFGGLRAEQLFSSALCAARLLRQRLPGPPDAPGAVFVLGGEGLRA
ELRAAGLRLAGDPSAGDGAAPRVRAVLVGYDEHFSFAKLREACAHLRDPECLLVATDRDP
WHPLSDGSRTPGTGSLAAAVETASGRQALVVGKPSPYMFECITENFSIDPARTLMVGDRL
ETDILFGHRCGMTTVLTLTGVSRLEEAQAYLAAGQHDLVPHYYVESIADLTEGLED
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q96GD0 |
| UniProtKB/Swiss-Prot Entry Name
| PLPP_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AY125047 |
| GeneCard ID
| PDXP |
| GenAtlas ID
| PDXP |
| HGNC ID
| HGNC:30259 |
| References |
|---|
| General References
| Not Available |
