| Identification |
|---|
| HMDB Protein ID
| CDBP03884 |
| Secondary Accession Numbers
| Not Available |
| Name
| Probable imidazolonepropionase |
| Description
| Not Available |
| Synonyms
|
- Amidohydrolase domain-containing protein 1
|
| Gene Name
| AMDHD1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides |
| Specific Function
| Not Available |
| GO Classification
|
| Biological Process |
| histidine catabolic process to glutamate and formamide |
| histidine catabolic process to glutamate and formate |
| histidine catabolic process |
| Cellular Component |
| cytosol |
| Component |
| cell part |
| intracellular part |
| cytoplasm |
| Function |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides |
| Molecular Function |
| imidazolonepropionase activity |
| metal ion binding |
| Process |
| glutamate metabolic process |
| metabolic process |
| cellular metabolic process |
| glutamine family amino acid metabolic process |
| histidine catabolic process to glutamate and formamide |
| cellular amino acid and derivative metabolic process |
| cellular amino acid metabolic process |
|
| Cellular Location
|
- Cytoplasmic
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Histidine Metabolism | 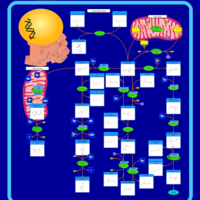 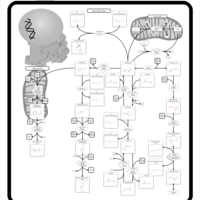 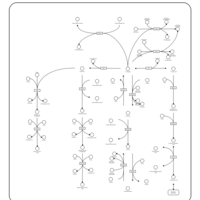 |  | | L-histidine degradation into L-glutamate | Not Available | Not Available | | Histidinemia | 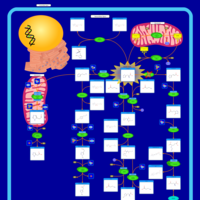   | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 12 |
| Locus
| 12q23.1 |
| SNPs
| AMDHD1 |
| Gene Sequence
|
>1281 bp
ATGGCAGGCGGCCACAGCCTCCTGCTGGAGAACGCGCAGCAAGTGGTGTTGGTGTGCGCC
CGCGGCGAGCGCTTCCTGGCGCGGGATGCGCTGCGCAGCCTGGCGGTGCTGGAAGGCGCC
AGCCTGGTGGTGGGCAAAGATGGATTTATAAAAGCTATTGGTCCTGCTGATGTTATTCAA
AGACAGTTTTCTGGAGAAACTTTTGAAGAAATAATTGACTGCTCTGGGAAATGTATCCTA
CCAGGTTTGGTGGATGCACACACACATCCAGTATGGGCTGGTGAAAGAGTTCACGAATTT
GCAATGAAGTTGGCAGGAGCCACCTACATGGAAATTCACCAGGCCGGAGGAGGGATCCAC
TTTACCGTGGAGCGCACGCGCCAAGCCACAGAGGAGGAGCTGTTCCGCTCCTTGCAGCAA
CGGCTCCAGTGCATGATGAGGGCTGGCACCACGCTGGTGGAGTGCAAGAGTGGATATGGC
CTCGACCTGGAGACCGAGCTCAAGATGCTGCGCGTGATTGAGCGCGCCCGGCGGGAGCTG
GACATCGGCATCTCGGCTACCTACTGCGGGGCTCATTCAGTGCCTAAAGGAAAAACTGCT
ACTGAAGCTGCTGATGACATCATCAATAACCACCTCCCAAAGCTGAAGGAACTTGGCAGA
AATGGGGAAATACACGTGGACAATATAGACGTATTTTGTGAGAAAGGTGTCTTTGATCTC
GATTCCACCAGAAGGATTCTTCAACGTGGAAAAGATATAGGGTTACAGATTAACTTCCAT
GGGGATGAACTCCACCCGATGAAGGCTGCTGAGCTTGGGGCTGAACTGGGAGCGCAGGCA
ATCAGCCACCTGGAAGAAGTGAGTGATGAAGGCATCGTTGCCATGGCAACGGCCAGGTGC
TCTGCCATCCTTCTGCCCACCACAGCCTACATGCTGAGACTGAAACAACCTCGAGCCAGG
AAGATGTTAGATGAAGGAGTAATAGTTGCTCTGGGAAGTGATTTCAACCCCAATGCATAT
TGCTTTTCAATGCCAATGGTCATGCATCTGGCCTGTGTAAACATGAGAATGTCCATGCCT
GAGGCCTTGGCCGCTGCCACCATCAATGCAGCTTATGCACTGGGAAAGTCTCACACACAC
GGATCGTTGGAAGTTGGCAAACAGGGAGATCTCATTATCATCAATTCATCCCGATGGGAG
CATTTGATTTACCAGTTCGGAGGCCATCATGAATTAATTGAATATGTTATAGCTAAAGGA
AAACTCATCTATAAAACATGA
|
| Protein Properties |
|---|
| Number of Residues
| 426 |
| Molecular Weight
| 46742.505 |
| Theoretical pI
| 6.611 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Probable imidazolonepropionase
MAGGHSLLLENAQQVVLVCARGERFLARDALRSLAVLEGASLVVGKDGFIKAIGPADVIQ
RQFSGETFEEIIDCSGKCILPGLVDAHTHPVWAGERVHEFAMKLAGATYMEIHQAGGGIH
FTVERTRQATEEELFRSLQQRLQCMMRAGTTLVECKSGYGLDLETELKMLRVIERARREL
DIGISATYCGAHSVPKGKTATEAADDIINNHLPKLKELGRNGEIHVDNIDVFCEKGVFDL
DSTRRILQRGKDIGLQINFHGDELHPMKAAELGAELGAQAISHLEEVSDEGIVAMATARC
SAILLPTTAYMLRLKQPRARKMLDEGVIVALGSDFNPNAYCFSMPMVMHLACVNMRMSMP
EALAAATINAAYALGKSHTHGSLEVGKQGDLIIINSSRWEHLIYQFGGHHELIEYVIAKG
KLIYKT
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q96NU7 |
| UniProtKB/Swiss-Prot Entry Name
| HUTI_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AK054617 |
| GeneCard ID
| AMDHD1 |
| GenAtlas ID
| AMDHD1 |
| HGNC ID
| HGNC:28577 |
| References |
|---|
| General References
| Not Available |
