| Identification |
|---|
| HMDB Protein ID
| CDBP02625 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ribonucleoside-diphosphate reductase subunit M2 B |
| Description
| Not Available |
| Synonyms
|
- TP53-inducible ribonucleotide reductase M2 B
- p53-inducible ribonucleotide reductase small subunit 2-like protein
- p53R2
|
| Gene Name
| RRM2B |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Plays a pivotal role in cell survival by repairing damaged DNA in a p53/TP53-dependent manner. Supplies deoxyribonucleotides for DNA repair in cells arrested at G1 or G2. Contains an iron-tyrosyl free radical center required for catalysis. Forms an active ribonucleotide reductase (RNR) complex with RRM1 which is expressed both in resting and proliferating cells in response to DNA damage.
|
| GO Classification
|
| Biological Process |
| mitochondrial DNA replication |
| negative regulation of apoptotic process |
| nucleobase-containing small molecule interconversion |
| DNA repair |
| response to oxidative stress |
| kidney development |
| deoxyribonucleotide biosynthetic process |
| deoxyribonucleoside diphosphate metabolic process |
| deoxyribonucleoside triphosphate metabolic process |
| renal system process |
| Cellular Component |
| nucleoplasm |
| ribonucleoside-diphosphate reductase complex |
| mitochondrion |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| oxidoreductase activity, acting on ch or ch2 groups |
| oxidoreductase activity, acting on ch or ch2 groups, disulfide as acceptor |
| ribonucleoside-diphosphate reductase activity |
| oxidoreductase activity |
| Molecular Function |
| transition metal ion binding |
| ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor |
| Process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| nucleoside diphosphate metabolic process |
| deoxyribonucleoside diphosphate metabolic process |
| oxidation reduction |
| metabolic process |
| nitrogen compound metabolic process |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Pyrimidine Metabolism |    |  | | Purine Metabolism |    |  | | DNA replication | Not Available | 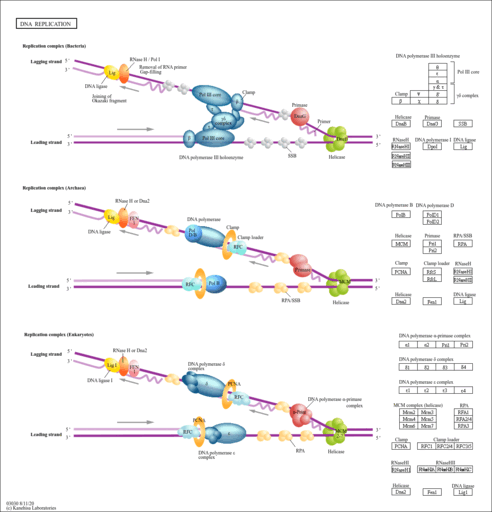 | | p53 signaling pathway | Not Available | 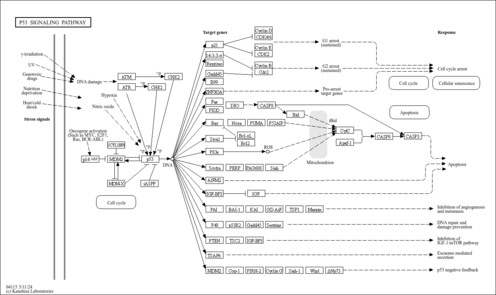 | | Beta Ureidopropionase Deficiency |  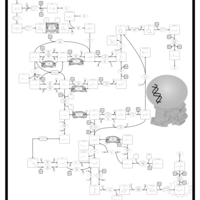 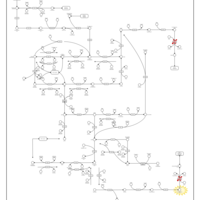 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 8 |
| Locus
| 8q23.1 |
| SNPs
| RRM2B |
| Gene Sequence
|
>1056 bp
ATGGGCGACCCGGAAAGGCCGGAAGCGGCCGGGCTGGATCAGGATGAGAGATCATCTTCA
GACACCAACGAAAGTGAAATAAAGTCAAATGAAGAGCCACTCCTAAGAAAGAGTTCTCGC
CGGTTTGTCATCTTTCCAATCCAGTACCCTGATATTTGGAAAATGTATAAACAGGCACAG
GCTTCCTTCTGGACAGCAGAAGAGGTCGACTTATCAAAGGATCTCCCTCACTGGAACAAG
CTTAAAGCAGATGAGAAGTACTTCATCTCTCACATCTTAGCCTTTTTTGCAGCCAGTGAT
GGAATTGTAAATGAAAATTTGGTGGAGCGCTTTAGTCAGGAGGTGCAGGTTCCAGAGGCT
CGCTGTTTCTATGGCTTTCAAATTCTCATCGAGAATGTTCACTCAGAGATGTACAGTTTG
CTGATAGACACTTACATCAGAGATCCCAAGAAAAGGGAATTTTTATTTAATGCAATTGAA
ACCATGCCCTATGTTAAGAAAAAAGCAGATTGGGCCTTGCGATGGATAGCAGATAGAAAA
TCTACTTTTGGGGAAAGAGTGGTGGCCTTTGCTGCTGTAGAAGGAGTTTTCTTCTCAGGA
TCTTTTGCTGCTATATTCTGGCTAAAGAAGAGAGGTCTTATGCCAGGACTCACTTTTTCC
AATGAACTCATCAGCAGAGATGAAGGACTTCACTGTGACTTTGCTTGCCTGATGTTCCAA
TACTTAGTAAATAAGCCTTCAGAAGAAAGGGTCAGGGAGATCATTGTTGATGCTGTCAAA
ATTGAGCAGGAGTTTTTAACAGAAGCCTTGCCAGTTGGCCTCATTGGAATGAATTGCATT
TTGATGAAACAGTACATTGAGTTTGTAGCTGACAGATTACTTGTGGAACTTGGATTCTCA
AAGGTTTTTCAGGCAGAAAATCCTTTTGATTTTATGGAAAACATTTCTTTAGAAGGAAAA
ACAAATTTCTTTGAGAAACGAGTTTCAGAGTATCAGCGTTTTGCAGTTATGGCAGAAACC
ACAGATAACGTCTTCACCTTGGATGCAGATTTTTAA
|
| Protein Properties |
|---|
| Number of Residues
| 351 |
| Molecular Weight
| 48786.6 |
| Theoretical pI
| 7.988 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ribonucleoside-diphosphate reductase subunit M2 B
MGDPERPEAAGLDQDERSSSDTNESEIKSNEEPLLRKSSRRFVIFPIQYPDIWKMYKQAQ
ASFWTAEEVDLSKDLPHWNKLKADEKYFISHILAFFAASDGIVNENLVERFSQEVQVPEA
RCFYGFQILIENVHSEMYSLLIDTYIRDPKKREFLFNAIETMPYVKKKADWALRWIADRK
STFGERVVAFAAVEGVFFSGSFAAIFWLKKRGLMPGLTFSNELISRDEGLHCDFACLMFQ
YLVNKPSEERVREIIVDAVKIEQEFLTEALPVGLIGMNCILMKQYIEFVADRLLVELGFS
KVFQAENPFDFMENISLEGKTNFFEKRVSEYQRFAVMAETTDNVFTLDADF
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q7LG56 |
| UniProtKB/Swiss-Prot Entry Name
| RIR2B_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AB036063 |
| GeneCard ID
| RRM2B |
| GenAtlas ID
| RRM2B |
| HGNC ID
| HGNC:17296 |
| References |
|---|
| General References
| Not Available |



