| Identification |
|---|
| HMDB Protein ID
| CDBP02622 |
| Secondary Accession Numbers
| Not Available |
| Name
| Peroxisomal NADH pyrophosphatase NUDT12 |
| Description
| Not Available |
| Synonyms
|
- Nucleoside diphosphate-linked moiety X motif 12
- Nudix motif 12
|
| Gene Name
| NUDT12 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity |
| Specific Function
| Hydrolyzes NAD(P)H to NMNH and AMP (2',5'-ADP), and diadenosine diphosphate to AMP. Has also activity towards NAD(P)(+), ADP-ribose and diadenosine triphosphate. May act to regulate the concentration of peroxisomal nicotinamide nucleotide cofactors required for oxidative metabolism in this organelle.
|
| GO Classification
|
| Biological Process |
| NAD catabolic process |
| NADP catabolic process |
| Cellular Component |
| nucleus |
| peroxisome |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| hydrolase activity |
| Molecular Function |
| NADH pyrophosphatase activity |
| NAD+ diphosphatase activity |
| metal ion binding |
|
| Cellular Location
|
- Peroxisome
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Nicotinate and Nicotinamide Metabolism | 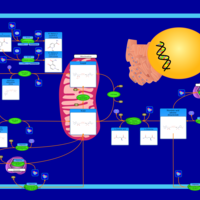   |  | | Peroxisome | Not Available | 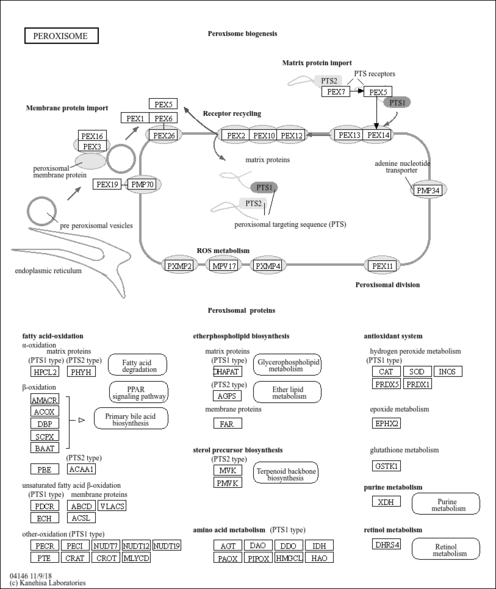 |
|
| Gene Properties |
|---|
| Chromosome Location
| 5 |
| Locus
| 5q21.2 |
| SNPs
| NUDT12 |
| Gene Sequence
|
>1389 bp
ATGTCTTCTGTAAAAAGAAGTCTGAAGCAAGAAATAGTTACTCAGTTTCACTGTTCAGCT
GCTGAAGGAGATATTGCCAAGTTAACAGGAATACTCAGTCATTCTCCATCTCTTCTCAAT
GAAACTTCTGAAAATGGCTGGACTGCTTTAATGTATGCGGCAAGGAATGGGCACCCAGAG
ATAGTCCAATTTCTGCTTGAGAAAGGGTGTGACAGATCAATTGTCAATAAATCAAGGCAG
ACTGCACTGGATATTGCTGTATTTTGGGGTTATAAGCATATAGCTAATTTACTAGCTACT
GCTAAAGGTGGGAAGAAGCCTTGGTTCCTAACGAATGAAGTGGAAGAATGTGAAAATTAT
TTTAGCAAAACACTACTGGACCGGAAAAGTGAAAAGAGGAATAATTCTGACTGGCTGCTA
GCTAAAGAAAGCCATCCAGCCACAGTTTTTATTCTTTTCTCAGATTTAAATCCCTTGGTT
ACTCTAGGTGGCAATAAAGAAAGTTTCCAACAGCCAGAAGTTAGGCTTTGTCAGCTGAAC
TACACAGATATAAAGGATTATTTGGCCCAGCCTGAGAAGATCACCTTGATTTTTCTTGGA
GTAGAACTTGAAATAAAAGACAAACTACTTAATTATGCTGGTGAAGTCCCGAGAGAGGAG
GAAGATGGATTGGTTGCCTGGTTTGCTCTAGGTATAGATCCTATTGCTGCTGAAGAATTC
AAGCAAAGACATGAAAATTGTTACTTTCTTCATCCTCCTATGCCAGCCCTTCTGCAATTG
AAAGAAAAAGAAGCTGGGGTTGTAGCTCAAGCAAGATCTGTTCTTGCCTGGCACAGTCGA
TACAAGTTTTGCCCAACCTGTGGAAATGCAACTAAAATTGAAGAAGGTGGCTATAAGAGA
CTATGTTTAAAAGAAGACTGTCCTAGTCTCAATGGCGTCCATAATACCTCATACCCAAGA
GTTGATCCAGTAGTAATCATGCAAGTTATTCATCCAGATGGGACCAAATGCCTTTTAGGC
AGGCAGAAAAGATTTCCCCCAGGCATGTTTACTTGCCTTGCTGGATTTATTGAGCCTGGA
GAGACAATAGAAGATGCTGTTAGGAGAGAAGTAGAAGAGGAAAGTGGAGTCAAAGTTGGC
CATGTTCAGTATGTTGCTTGTCAACCATGGCCAATGCCTTCCTCCTTAATGATTGGTTGC
TTAGCTCTAGCAGTGTCTACAGAAATTAAAGTTGACAAGAATGAAATAGAGGATGCCCGC
TGGTTCACTAGAGAACAGGTCCTGGATGTTCTGACCAAAGGGAAGCAGCAGGCATTCTTT
GTGCCACCAAGCCGAGCTATTGCACATCAATTAATCAAACACTGGATTAGAATAAATCCT
AATCTCTAA
|
| Protein Properties |
|---|
| Number of Residues
| 462 |
| Molecular Weight
| 52075.475 |
| Theoretical pI
| 6.829 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Peroxisomal NADH pyrophosphatase NUDT12
MSSVKRSLKQEIVTQFHCSAAEGDIAKLTGILSHSPSLLNETSENGWTALMYAARNGHPE
IVQFLLEKGCDRSIVNKSRQTALDIAVFWGYKHIANLLATAKGGKKPWFLTNEVEECENY
FSKTLLDRKSEKRNNSDWLLAKESHPATVFILFSDLNPLVTLGGNKESFQQPEVRLCQLN
YTDIKDYLAQPEKITLIFLGVELEIKDKLLNYAGEVPREEEDGLVAWFALGIDPIAAEEF
KQRHENCYFLHPPMPALLQLKEKEAGVVAQARSVLAWHSRYKFCPTCGNATKIEEGGYKR
LCLKEDCPSLNGVHNTSYPRVDPVVIMQVIHPDGTKCLLGRQKRFPPGMFTCLAGFIEPG
ETIEDAVRREVEEESGVKVGHVQYVACQPWPMPSSLMIGCLALAVSTEIKVDKNEIEDAR
WFTREQVLDVLTKGKQQAFFVPPSRAIAHQLIKHWIRINPNL
|
| External Links |
|---|
| GenBank ID Protein
| 193783663 |
| UniProtKB/Swiss-Prot ID
| Q9BQG2 |
| UniProtKB/Swiss-Prot Entry Name
| NUD12_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AK098066 |
| GeneCard ID
| NUDT12 |
| GenAtlas ID
| NUDT12 |
| HGNC ID
| HGNC:18826 |
| References |
|---|
| General References
| Not Available |

