| Identification |
|---|
| HMDB Protein ID
| CDBP02231 |
| Secondary Accession Numbers
| Not Available |
| Name
| Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- BCKAD-E2
- BCKADE2
- Branched-chain alpha-keto acid dehydrogenase complex component E2
- Dihydrolipoamide acetyltransferase component of branched-chain alpha-keto acid dehydrogenase complex
- Dihydrolipoamide branched chain transacylase
- Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase
|
| Gene Name
| DBT |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acyltransferase activity |
| Specific Function
| The branched-chain alpha-keto dehydrogenase complex catalyzes the overall conversion of alpha-keto acids to acyl-CoA and CO(2). It contains multiple copies of three enzymatic components: branched-chain alpha-keto acid decarboxylase (E1), lipoamide acyltransferase (E2) and lipoamide dehydrogenase (E3).
|
| GO Classification
|
| Biological Process |
| branched-chain amino acid catabolic process |
| cellular nitrogen compound metabolic process |
| fatty-acyl-CoA biosynthetic process |
| Cellular Component |
| mitochondrial nucleoid |
| microtubule cytoskeleton |
| mitochondrial alpha-ketoglutarate dehydrogenase complex |
| Function |
| binding |
| catalytic activity |
| transferase activity |
| transferase activity, transferring acyl groups |
| transferase activity, transferring acyl groups other than amino-acyl groups |
| acyltransferase activity |
| protein binding |
| cofactor binding |
| dihydrolipoyllysine-residue (2-methylpropanoyl)transferase activity |
| Molecular Function |
| dihydrolipoyllysine-residue (2-methylpropanoyl)transferase activity |
| cofactor binding |
| Process |
| fatty-acyl-coa biosynthetic process |
| cofactor metabolic process |
| coenzyme metabolic process |
| acyl-coa metabolic process |
| metabolic process |
| cellular metabolic process |
| fatty-acyl-coa metabolic process |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Threonine and 2-Oxobutanoate Degradation | 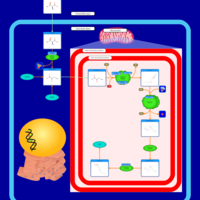 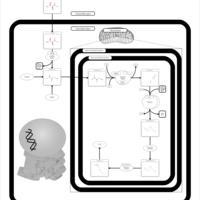 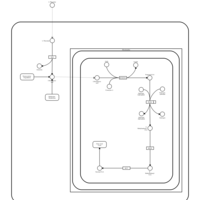 | Not Available | | Valine, Leucine and Isoleucine Degradation |  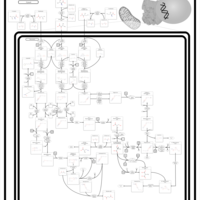 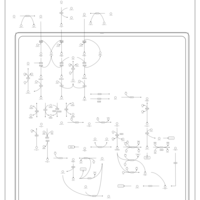 |  | | Beta-Ketothiolase Deficiency |  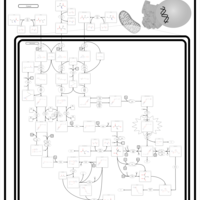 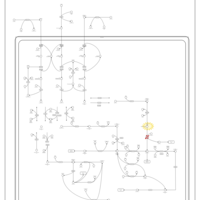 | Not Available | | 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency | 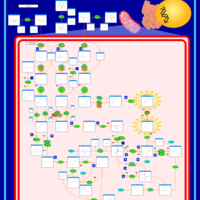 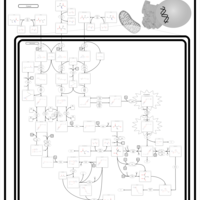 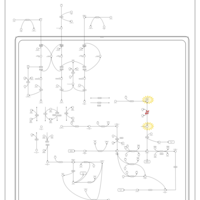 | Not Available | | Propionic Acidemia | 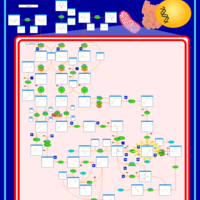 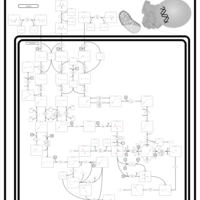 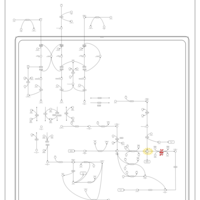 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1p31 |
| SNPs
| DBT |
| Gene Sequence
|
>1449 bp
ATGGCTGCAGTCCGTATGCTGAGAACCTGGAGCAGGAATGCGGGGAAGCTGATTTGTGTT
CGCTATTTTCAAACATGTGGTAATGTTCATGTTTTGAAGCCAAATTATGTGTGTTTCTTT
GGTTATCCTTCATTCAAGTATAGTCATCCACATCACTTCCTGAAAACAACTGCTGCTCTC
CGTGGACAGGTTGTTCAGTTCAAGCTCTCAGACATTGGAGAAGGGATTAGAGAAGTAACT
GTTAAAGAATGGTATGTAAAAGAAGGAGATACAGTGTCTCAGTTTGATAGCATCTGTGAA
GTTCAAAGTGATAAAGCTTCTGTTACCATCACTAGTCGTTATGATGGAGTCATTAAAAAA
CTCTATTATAATCTAGACGATATTGCCTATGTGGGGAAGCCATTAGTAGACATAGAAACG
GAAGCTTTAAAAGATTCAGAAGAAGATGTTGTTGAAACTCCTGCAGTGTCTCATGATGAA
CATACACACCAAGAGATAAAGGGCCGAAAAACACTGGCAACTCCTGCAGTTCGCCGTCTG
GCAATGGAAAACAATATTAAGCTGAGTGAAGTTGTTGGCTCAGGAAAAGATGGCAGAATA
CTTAAAGAAGATATCCTCAACTATTTGGAAAAGCAGACAGGAGCTATATTGCCTCCTTCA
CCCAAAGTTGAAATTATGCCACCTCCACCAAAGCCAAAAGACATGACTGTTCCTATACTA
GTATCAAAACCTCCGGTATTCACAGGCAAAGACAAAACAGAACCCATAAAAGGCTTTCAA
AAAGCAATGGTCAAGACTATGTCTGCAGCCCTGAAGATACCTCATTTTGGTTATTGTGAT
GAGATTGACCTTACTGAACTGGTTAAGCTCCGAGAAGAATTAAAACCCATTGCATTTGCT
CGTGGAATTAAACTCTCCTTTATGCCTTTCTTCTTAAAGGCTGCTTCCTTGGGATTACTA
CAGTTTCCTATCCTTAACGCTTCTGTGGATGAAAACTGCCAGAATATAACATATAAGGCT
TCTCATAACATTGGGATAGCAATGGATACTGAGCAGGGTTTGATTGTCCCTAATGTGAAA
AATGTTCAGATCTGCTCTATATTTGACATCGCCACTGAACTGAACCGCCTCCAGAAATTG
GGCTCTGTGGGTCAGCTCAGCACCACTGATCTTACAGGAGGAACATTTACTCTTTCCAAC
ATTGGATCAATTGGTGGTACCTTTGCCAAACCAGTGATAATGCCACCTGAAGTAGCCATT
GGGGCCCTTGGATCAATTAAGGCCATTCCCCGATTTAACCAGAAAGGAGAAGTATATAAG
GCACAGATAATGAATGTGAGCTGGTCAGCTGATCACAGAGTTATTGATGGTGCTACAATG
TCACGCTTCTCCAATTTGTGGAAATCCTATTTAGAAAACCCAGCTTTTATGCTACTAGAT
CTGAAATGA
|
| Protein Properties |
|---|
| Number of Residues
| 482 |
| Molecular Weight
| 53486.635 |
| Theoretical pI
| 8.518 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial
MAAVRMLRTWSRNAGKLICVRYFQTCGNVHVLKPNYVCFFGYPSFKYSHPHHFLKTTAAL
RGQVVQFKLSDIGEGIREVTVKEWYVKEGDTVSQFDSICEVQSDKASVTITSRYDGVIKK
LYYNLDDIAYVGKPLVDIETEALKDSEEDVVETPAVSHDEHTHQEIKGRKTLATPAVRRL
AMENNIKLSEVVGSGKDGRILKEDILNYLEKQTGAILPPSPKVEIMPPPPKPKDMTVPIL
VSKPPVFTGKDKTEPIKGFQKAMVKTMSAALKIPHFGYCDEIDLTELVKLREELKPIAFA
RGIKLSFMPFFLKAASLGLLQFPILNASVDENCQNITYKASHNIGIAMDTEQGLIVPNVK
NVQICSIFDIATELNRLQKLGSVGQLSTTDLTGGTFTLSNIGSIGGTFAKPVIMPPEVAI
GALGSIKAIPRFNQKGEVYKAQIMNVSWSADHRVIDGATMSRFSNLWKSYLENPAFMLLD
LK
|
| External Links |
|---|
| GenBank ID Protein
| 189053756 |
| UniProtKB/Swiss-Prot ID
| P11182 |
| UniProtKB/Swiss-Prot Entry Name
| ODB2_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AK313191 |
| GeneCard ID
| DBT |
| GenAtlas ID
| DBT |
| HGNC ID
| HGNC:2698 |
| References |
|---|
| General References
| Not Available |
