| Identification |
|---|
| HMDB Protein ID
| CDBP01609 |
| Secondary Accession Numbers
| Not Available |
| Name
| Bifunctional epoxide hydrolase 2 |
| Description
| Not Available |
| Synonyms
|
- CEH
- Cytosolic epoxide hydrolase
- Epoxide hydratase
- SEH
- Soluble epoxide hydrolase
|
| Gene Name
| EPHX2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| Bifunctional enzyme. The C-terminal domain has epoxide hydrolase activity and acts on epoxides (alkene oxides, oxiranes) and arene oxides. Plays a role in xenobiotic metabolism by degrading potentially toxic epoxides. Also determines steady-state levels of physiological mediators. The N-terminal domain has lipid phosphatase activity, with the highest activity towards threo-9,10-phosphonooxy-hydroxy-octadecanoic acid, followed by erythro-9,10-phosphonooxy-hydroxy-octadecanoic acid, 12-phosphonooxy-octadec-9Z-enoic acid, 12-phosphonooxy-octadec-9E-enoic acid, and p-nitrophenyl phospate.
|
| GO Classification
|
| Biological Process |
| regulation of cholesterol metabolic process |
| drug metabolic process |
| phospholipid dephosphorylation |
| stilbene catabolic process |
| inflammatory response |
| reactive oxygen species metabolic process |
| positive regulation of vasodilation |
| xenobiotic metabolic process |
| cellular calcium ion homeostasis |
| cholesterol homeostasis |
| regulation of blood pressure |
| response to toxin |
| positive regulation of gene expression |
| Cellular Component |
| cytosol |
| focal adhesion |
| nucleolus |
| Golgi apparatus |
| peroxisome |
| Function |
| catalytic activity |
| hydrolase activity |
| Molecular Function |
| magnesium ion binding |
| epoxide hydrolase activity |
| 10-hydroxy-9-(phosphonooxy)octadecanoate phosphatase activity |
| lipid phosphatase activity |
| toxin binding |
| 4-nitrophenylphosphatase activity |
| protein homodimerization activity |
| Process |
| metabolic process |
|
| Cellular Location
|
- Cytoplasm
- Peroxisome
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Peroxisome | Not Available | 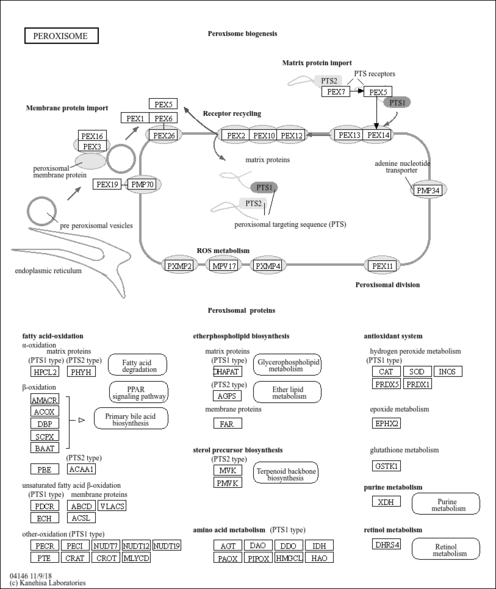 | | Arachidonic Acid Metabolism |    |  | | Leukotriene C4 Synthesis Deficiency |    | Not Available | | Piroxicam Action Pathway |   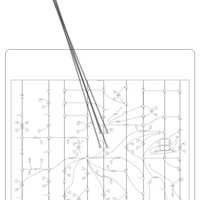 | Not Available | | Acetylsalicylic Acid Action Pathway |  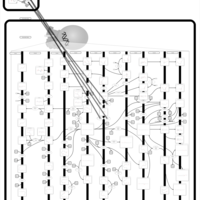 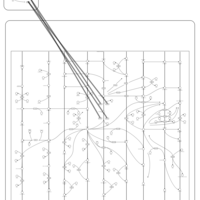 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 8 |
| Locus
| 8p21 |
| SNPs
| EPHX2 |
| Gene Sequence
|
>1668 bp
ATGACGCTGCGCGCGGCCGTCTTCGACCTTGACGGGGTGCTGGCGCTGCCAGCGGTGTTC
GGCGTCCTCGGCCGCACGGAGGAGGCCCTGGCGCTGCCCAGAGGACTTCTGAATGATGCT
TTCCAGAAAGGGGGACCAGAGGGTGCCACTACCCGGCTTATGAAAGGAGAGATCACACTT
TCCCAGTGGATACCACTCATGGAAGAAAACTGCAGGAAGTGCTCCGAGACCGCTAAAGTC
TGCCTCCCCAAGAATTTCTCCATAAAAGAAATCTTTGACAAGGCGATTTCAGCCAGAAAG
ATCAACCGCCCCATGCTCCAGGCAGCTCTCATGCTCAGGAAGAAAGGATTCACTACTGCC
ATCCTCACCAACACCTGGCTGGACGACCGTGCTGAGAGAGATGGCCTGGCCCAGCTGATG
TGTGAGCTGAAGATGCACTTTGACTTCCTGATAGAGTCGTGTCAGGTGGGAATGGTCAAA
CCTGAACCTCAGATCTACAAGTTTCTGCTGGACACCCTGAAGGCCAGCCCCAGTGAGGTC
GTTTTTTTGGATGACATCGGGGCTAATCTGAAGCCAGCCCGTGACTTGGGAATGGTCACC
ATCCTGGTCCAGGACACTGACACGGCCCTGAAAGAACTGGAGAAAGTGACCGGAATCCAG
CTTCTCAATACCCCGGCCCCTCTGCCGACCTCTTGCAATCCAAGTGACATGAGCCATGGG
TACGTGACAGTAAAGCCCAGGGTCCGTCTGCATTTTGTGGAGCTGGGCTCCGGCCCTGCT
GTGTGCCTCTGCCATGGATTTCCCGAGAGTTGGTATTCTTGGAGGTACCAGATCCCTGCT
CTGGCCCAGGCAGGTTACCGGGTCCTAGCTATGGACATGAAAGGCTATGGAGAGTCATCT
GCTCCTCCCGAAATAGAAGAATATTGCATGGAAGTGTTATGTAAGGAGATGGTAACCTTC
CTGGATAAACTGGGCCTCTCTCAAGCAGTGTTCATTGGCCATGACTGGGGTGGCATGCTG
GTGTGGTACATGGCTCTCTTCTACCCCGAGAGAGTGAGGGCGGTGGCCAGTTTGAATACT
CCCTTCATACCAGCAAATCCCAACATGTCCCCTTTGGAGAGTATCAAAGCCAACCCAGTA
TTTGATTACCAGCTCTACTTCCAAGAACCAGGAGTGGCTGAGGCTGAACTGGAACAGAAC
CTGAGTCGGACTTTCAAAAGCCTCTTCAGAGCAAGCGATGAGAGTGTTTTATCCATGCAT
AAAGTCTGTGAAGCGGGAGGACTTTTTGTAAATAGCCCAGAAGAGCCCAGCCTCAGCAGG
ATGGTCACTGAGGAGGAAATCCAGTTCTATGTGCAGCAGTTCAAGAAGTCTGGTTTCAGA
GGTCCTCTAAACTGGTACCGAAACATGGAAAGGAACTGGAAGTGGGCTTGCAAAAGCTTG
GGACGGAAGATCCTGATTCCGGCCCTGATGGTCACGGCGGAGAAGGACTTCGTGCTCGTT
CCTCAGATGTCCCAGCACATGGAGGACTGGATTCCCCACCTGAAAAGGGGACACATTGAG
GACTGTGGGCACTGGACACAGATGGACAAGCCAACCGAGGTGAATCAGATCCTCATTAAG
TGGCTGGATTCTGATGCCCGGAACCCACCGGTGGTCTCAAAGATGTAG
|
| Protein Properties |
|---|
| Number of Residues
| 555 |
| Molecular Weight
| 62615.22 |
| Theoretical pI
| 6.281 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Epoxide hydrolase 2
MTLRAAVFDLDGVLALPAVFGVLGRTEEALALPRGLLNDAFQKGGPEGATTRLMKGEITL
SQWIPLMEENCRKCSETAKVCLPKNFSIKEIFDKAISARKINRPMLQAALMLRKKGFTTA
ILTNTWLDDRAERDGLAQLMCELKMHFDFLIESCQVGMVKPEPQIYKFLLDTLKASPSEV
VFLDDIGANLKPARDLGMVTILVQDTDTALKELEKVTGIQLLNTPAPLPTSCNPSDMSHG
YVTVKPRVRLHFVELGSGPAVCLCHGFPESWYSWRYQIPALAQAGYRVLAMDMKGYGESS
APPEIEEYCMEVLCKEMVTFLDKLGLSQAVFIGHDWGGMLVWYMALFYPERVRAVASLNT
PFIPANPNMSPLESIKANPVFDYQLYFQEPGVAEAELEQNLSRTFKSLFRASDESVLSMH
KVCEAGGLFVNSPEEPSLSRMVTEEEIQFYVQQFKKSGFRGPLNWYRNMERNWKWACKSL
GRKILIPALMVTAEKDFVLVPQMSQHMEDWIPHLKRGHIEDCGHWTQMDKPTEVNQILIK
WLDSDARNPPVVSKM
|
| External Links |
|---|
| GenBank ID Protein
| 10197680 |
| UniProtKB/Swiss-Prot ID
| P34913 |
| UniProtKB/Swiss-Prot Entry Name
| HYES_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF233334 |
| GeneCard ID
| EPHX2 |
| GenAtlas ID
| EPHX2 |
| HGNC ID
| HGNC:3402 |
| References |
|---|
| General References
| Not Available |

