| Identification |
|---|
| HMDB Protein ID
| CDBP01492 |
| Secondary Accession Numbers
| Not Available |
| Name
| Phosphomannomutase 1 |
| Description
| Not Available |
| Synonyms
|
- PMM 1
- PMMH-22
|
| Gene Name
| PMM1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in metal ion binding |
| Specific Function
| Involved in the synthesis of the GDP-mannose and dolichol-phosphate-mannose required for a number of critical mannosyl transfer reactions. In addition, may be responsible for the degradation of glucose-1,6-bisphosphate in ischemic brain.
|
| GO Classification
|
| Biological Process |
| dolichol-linked oligosaccharide biosynthetic process |
| post-translational protein modification |
| protein N-linked glycosylation via asparagine |
| mannose biosynthetic process |
| mannose metabolic process |
| GDP-mannose biosynthetic process |
| Cellular Component |
| cytosol |
| neuronal cell body |
| Component |
| cell part |
| intracellular part |
| cytoplasm |
| Function |
| intramolecular transferase activity, phosphotransferases |
| catalytic activity |
| isomerase activity |
| phosphomannomutase activity |
| intramolecular transferase activity |
| Molecular Function |
| metal ion binding |
| phosphomannomutase activity |
| Process |
| metabolic process |
| mannose metabolic process |
| small molecule metabolic process |
| alcohol metabolic process |
| monosaccharide metabolic process |
| hexose metabolic process |
| mannose biosynthetic process |
|
| Cellular Location
|
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | GDP-alpha-D-mannose biosynthesis | Not Available | Not Available | | Fructose and mannose metabolism | Not Available |  | | Amino sugar and nucleotide sugar metabolism | Not Available |  | | Fructose and Mannose Degradation | 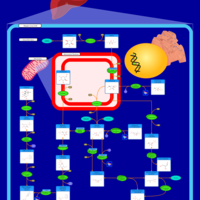 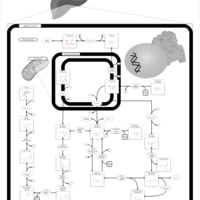 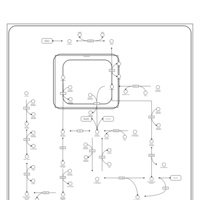 |  | | Fructosuria | 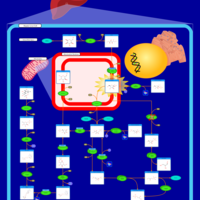   | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 22 |
| Locus
| 22q13.2 |
| SNPs
| PMM1 |
| Gene Sequence
|
>789 bp
ATGGCAGTCACCGCCCAGGCAGCCCGCAGGAAGGAGCGCGTCCTCTGCCTGTTTGACGTG
GACGGGACCCTCACGCCGGCTCGCCAGAAAATTGACCCTGAGGTGGCCGCCTTCCTGCAG
AAGCTACGAAGTAGAGTGCAGATCGGTGTGGTGGGCGGCTCTGACTACTGTAAGATCGCT
GAGCAGCTGGGTGACGGGGATGAAGTCATTGAGAAGTTTGATTATGTGTTTGCCGAGAAC
GGGACGGTGCAGTATAAGCACGGACGACTGCTCTCCAAGCAGACCATCCAGAACCACCTG
GGGGAGGAGCTGCTGCAGGACTTGATCAACTTCTGCCTCAGCTACATGGCCCTGCTCAGG
CTGCCCAAGAAGCGTGGAACCTTCATCGAGTTCCGGAATGGCATGCTGAACATCTCGCCC
ATCGGCCGGAGCTGCACCCTGGAGGAGAGGATCGAGTTCTCCGAACTGGACAAGAAAGAG
AAGATCCGGGAGAAGTTCGTGGAAGCCCTGAAAACAGAGTTTGCTGGCAAAGGGCTGAGG
TTCTCTCGAGGAGGCATGATCAGCTTTGACGTCTTCCCCGAGGGCTGGGACAAGCGCTAC
TGCCTGGATAGCCTGGACCAGGACAGCTTCGACACCATCCACTTCTTTGGGAACGAGACT
AGCCCTGGTGGGAACGACTTTGAGATCTTTGCCGACCCCCGGACTGTTGGCCACAGCGTG
GTGTCTCCTCAGGACACGGTGCAGCGATGCCGGGAGATTTTCTTCCCAGAGACAGCTCAT
GAGGCGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 262 |
| Molecular Weight
| 29746.545 |
| Theoretical pI
| 5.747 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Phosphomannomutase 1
MAVTAQAARRKERVLCLFDVDGTLTPARQKIDPEVAAFLQKLRSRVQIGVVGGSDYCKIA
EQLGDGDEVIEKFDYVFAENGTVQYKHGRLLSKQTIQNHLGEELLQDLINFCLSYMALLR
LPKKRGTFIEFRNGMLNISPIGRSCTLEERIEFSELDKKEKIREKFVEALKTEFAGKGLR
FSRGGMISFDVFPEGWDKRYCLDSLDQDSFDTIHFFGNETSPGGNDFEIFADPRTVGHSV
VSPQDTVQRCREIFFPETAHEA
|
| External Links |
|---|
| GenBank ID Protein
| 5304855 |
| UniProtKB/Swiss-Prot ID
| Q92871 |
| UniProtKB/Swiss-Prot Entry Name
| PMM1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AL023553 |
| GeneCard ID
| PMM1 |
| GenAtlas ID
| Not Available |
| HGNC ID
| HGNC:9114 |
| References |
|---|
| General References
| Not Available |

