| Identification |
|---|
| HMDB Protein ID
| CDBP01470 |
| Secondary Accession Numbers
| Not Available |
| Name
| Catalase |
| Description
| Not Available |
| Synonyms
|
Not Available
|
| Gene Name
| CAT |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalase activity |
| Specific Function
| Occurs in almost all aerobically respiring organisms and serves to protect cells from the toxic effects of hydrogen peroxide. Promotes growth of cells including T-cells, B-cells, myeloid leukemia cells, melanoma cells, mastocytoma cells and normal and transformed fibroblast cells.
|
| GO Classification
|
| Biological Process |
| cellular response to growth factor stimulus |
| hemoglobin metabolic process |
| hydrogen peroxide catabolic process |
| menopause |
| positive regulation of cell division |
| positive regulation of NF-kappaB transcription factor activity |
| positive regulation of phosphatidylinositol 3-kinase cascade |
| response to hyperoxia |
| triglyceride metabolic process |
| UV protection |
| purine nucleobase metabolic process |
| response to hypoxia |
| purine nucleotide catabolic process |
| response to vitamin E |
| negative regulation of apoptotic process |
| cholesterol metabolic process |
| negative regulation of NF-kappaB transcription factor activity |
| protein homotetramerization |
| aerobic respiration |
| Cellular Component |
| lysosome |
| peroxisomal matrix |
| peroxisomal membrane |
| mitochondrial intermembrane space |
| cytosol |
| endoplasmic reticulum |
| plasma membrane |
| Golgi apparatus |
| Function |
| peroxidase activity |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| transition metal ion binding |
| iron ion binding |
| antioxidant activity |
| heme binding |
| catalase activity |
| Molecular Function |
| heme binding |
| protein homodimerization activity |
| aminoacylase activity |
| catalase activity |
| metal ion binding |
| NADP binding |
| Process |
| response to stimulus |
| response to stress |
| response to oxidative stress |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Peroxisome
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glyoxylate and dicarboxylate metabolism | Not Available | 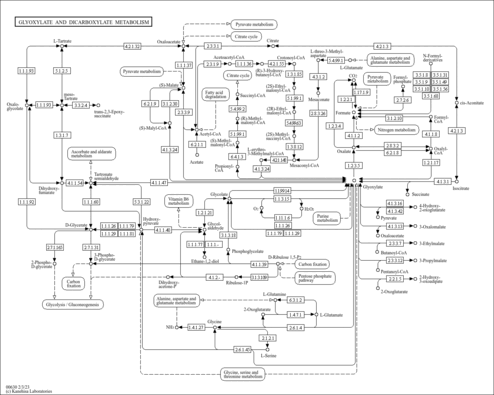 | | Peroxisome | Not Available | 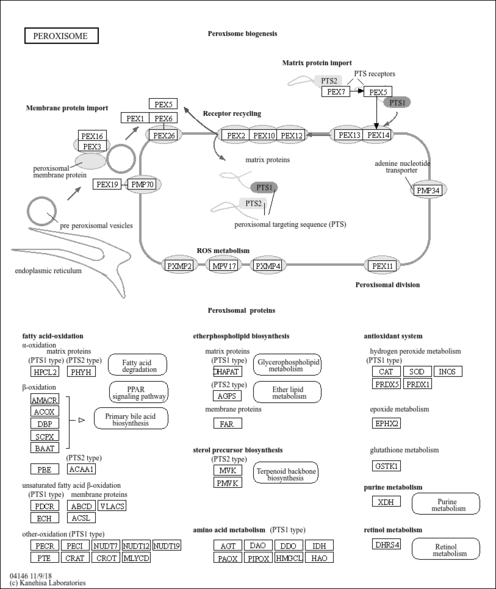 | | Amyotrophic lateral sclerosis (ALS) | Not Available | 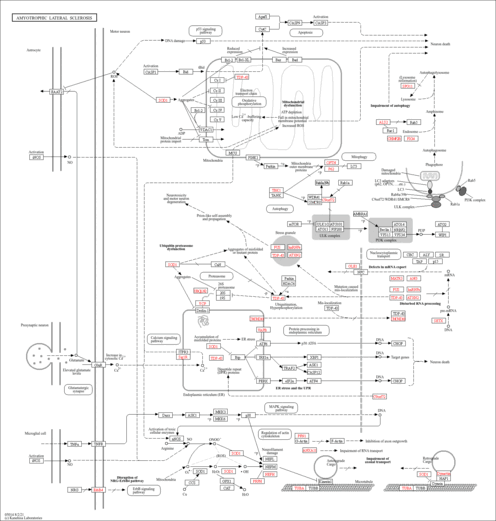 | | Degradation of Superoxides |    | Not Available | | Ethanol Degradation |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 11 |
| Locus
| 11p13 |
| SNPs
| CAT |
| Gene Sequence
|
>1584 bp
ATGGCTGACAGCCGGGATCCCGCCAGCGACCAGATGCAGCACTGGAAGGAGCAGCGGGCC
GCGCAGAAAGCTGATGTCCTGACCACTGGAGCTGGTAACCCAGTAGGAGACAAACTTAAT
GTTATTACAGTAGGGCCCCGTGGGCCCCTTCTTGTTCAGGATGTGGTTTTCACTGATGAA
ATGGCTCATTTTGACCGAGAGAGAATTCCTGAGAGAGTTGTGCATGCTAAAGGAGCAGGG
GCCTTTGGCTACTTTGAGGTCACACATGACATTACCAAATACTCCAAGGCAAAGGTATTT
GAGCATATTGGAAAGAAGACTCCCATCGCAGTTCGGTTCTCCACTGTTGCTGGAGAATCG
GGTTCAGCTGACACAGTTCGGGACCCTCGTGGGTTTGCAGTGAAATTTTACACAGAAGAT
GGTAACTGGGATCTCGTTGGAAATAACACCCCCATTTTCTTCATCAGGGATCCCATATTG
TTTCCATCTTTTATCCACAGCCAAAAGAGAAATCCTCAGACACATCTGAAGGATCCGGAC
ATGGTCTGGGACTTCTGGAGCCTACGTCCTGAGTCTCTGCATCAGGTTTCTTTCTTGTTC
AGTGATCGGGGGATTCCAGATGGACATCGCCACATGAATGGATATGGATCACATACTTTC
AAGCTGGTTAATGCAAATGGGGAGGCAGTTTATTGCAAATTCCATTATAAGACTGACCAG
GGCATCAAAAACCTTTCTGTTGAAGATGCGGCGAGACTTTCCCAGGAAGATCCTGACTAT
GGCATCCGGGATCTTTTTAACGCCATTGCCACAGGAAAGTACCCCTCCTGGACTTTTTAC
ATCCAGGTCATGACATTTAATCAGGCAGAAACTTTTCCATTTAATCCATTCGATCTCACC
AAGGTTTGGCCTCACAAGGACTACCCTCTCATCCCAGTTGGTAAACTGGTCTTAAACCGG
AATCCAGTTAATTACTTTGCTGAGGTTGAACAGATAGCCTTCGACCCAAGCAACATGCCA
CCTGGCATTGAGGCCAGTCCTGACAAAATGCTTCAGGGCCGCCTTTTTGCCTATCCTGAC
ACTCACCGCCATCGCCTGGGACCCAATTATCTTCATATACCTGTGAACTGTCCCTACCGT
GCTCGAGTGGCCAACTACCAGCGTGATGGCCCGATGTGCATGCAGGACAATCAGGGTGGT
GCTCCAAATTACTACCCCAACAGCTTTGGTGCTCCGGAACAACAGCCTTCTGCCCTGGAG
CACAGCATCCAATATTCTGGAGAAGTGCGGAGATTCAACACTGCCAATGATGATAACGTT
ACTCAGGTGCGGGCATTCTATGTGAACGTGCTGAATGAGGAACAGAGGAAACGTCTGTGT
GAGAACATTGCCGGCCACCTGAAGGATGCACAAATTTTCATCCAGAAGAAAGCGGTCAAG
AACTTCACTGAGGTCCACCCTGACTACGGGAGCCACATCCAGGCTCTTCTGGACAAGTAC
AATGCTGAGAAGCCTAAGAATGCGATTCACACCTTTGTGCAGTCCGGATCTCACTTGGCG
GCAAGGGAGAAGGCAAATCTGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 527 |
| Molecular Weight
| 59755.82 |
| Theoretical pI
| 7.384 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Catalase
MADSRDPASDQMQHWKEQRAAQKADVLTTGAGNPVGDKLNVITVGPRGPLLVQDVVFTDE
MAHFDRERIPERVVHAKGAGAFGYFEVTHDITKYSKAKVFEHIGKKTPIAVRFSTVAGES
GSADTVRDPRGFAVKFYTEDGNWDLVGNNTPIFFIRDPILFPSFIHSQKRNPQTHLKDPD
MVWDFWSLRPESLHQVSFLFSDRGIPDGHRHMNGYGSHTFKLVNANGEAVYCKFHYKTDQ
GIKNLSVEDAARLSQEDPDYGIRDLFNAIATGKYPSWTFYIQVMTFNQAETFPFNPFDLT
KVWPHKDYPLIPVGKLVLNRNPVNYFAEVEQIAFDPSNMPPGIEASPDKMLQGRLFAYPD
THRHRLGPNYLHIPVNCPYRARVANYQRDGPMCMQDNQGGAPNYYPNSFGAPEQQPSALE
HSIQYSGEVRRFNTANDDNVTQVRAFYVNVLNEEQRKRLCENIAGHLKDAQIFIQKKAVK
NFTEVHPDYGSHIQALLDKYNAEKPKNAIHTFVQSGSHLAAREKANL
|
| External Links |
|---|
| GenBank ID Protein
| 29721 |
| UniProtKB/Swiss-Prot ID
| P04040 |
| UniProtKB/Swiss-Prot Entry Name
| CATA_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| X04076 |
| GeneCard ID
| CAT |
| GenAtlas ID
| CAT |
| HGNC ID
| HGNC:1516 |
| References |
|---|
| General References
| Not Available |


