| Identification |
|---|
| HMDB Protein ID
| CDBP01464 |
| Secondary Accession Numbers
| Not Available |
| Name
| Prostaglandin-H2 D-isomerase |
| Description
| Not Available |
| Synonyms
|
- Beta-trace protein
- Cerebrin-28
- Glutathione-independent PGD synthase
- Lipocalin-type prostaglandin-D synthase
- PGD2 synthase
- PGDS
- PGDS2
- Prostaglandin-D2 synthase
|
| Gene Name
| PTGDS |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in binding |
| Specific Function
| Catalyzes the conversion of PGH2 to PGD2, a prostaglandin involved in smooth muscle contraction/relaxation and a potent inhibitor of platelet aggregation. Involved in a variety of CNS functions, such as sedation, NREM sleep and PGE2-induced allodynia, and may have an anti-apoptotic role in oligodendrocytes. Binds small non-substrate lipophilic molecules, including biliverdin, bilirubin, retinal, retinoic acid and thyroid hormone, and may act as a scavenger for harmful hydrophopic molecules and as a secretory retinoid and thyroid hormone transporter. Possibly involved in development and maintenance of the blood-brain, blood-retina, blood-aqueous humor and blood-testis barrier. It is likely to play important roles in both maturation and maintenance of the central nervous system and male reproductive system.
|
| GO Classification
|
| Biological Process |
| regulation of circadian sleep/wake cycle, sleep |
| response to glucocorticoid stimulus |
| prostaglandin biosynthetic process |
| Cellular Component |
| Golgi apparatus |
| perinuclear region of cytoplasm |
| nuclear membrane |
| extracellular space |
| rough endoplasmic reticulum |
| Function |
| transporter activity |
| binding |
| Molecular Function |
| retinoid binding |
| small molecule binding |
| fatty acid binding |
| transporter activity |
| prostaglandin-D synthase activity |
| Process |
| metabolic process |
| primary metabolic process |
| establishment of localization |
| transport |
| lipid metabolic process |
|
| Cellular Location
|
- Cytoplasm
- perinuclear region
- Secreted
- Golgi apparatus
- Nucleus membrane
- Rough endoplasmic reticulum
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Arachidonic Acid Metabolism | 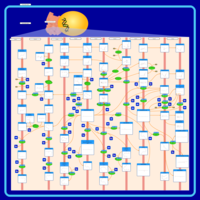  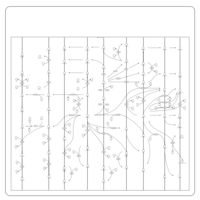 |  | | Leukotriene C4 Synthesis Deficiency | 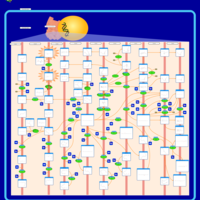 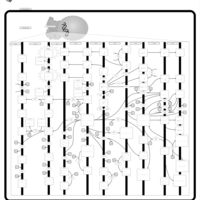  | Not Available | | Piroxicam Action Pathway |    | Not Available | | Acetylsalicylic Acid Action Pathway |    | Not Available | | Etodolac Action Pathway |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 9 |
| Locus
| 9q34.2-q34.3 |
| SNPs
| PTGDS |
| Gene Sequence
|
>573 bp
ATGGCTACTCATCACACGCTGTGGATGGGACTGGCCCTGCTGGGGGTGCTGGGCGACCTG
CAGGCAGCACCGGAGGCCCAGGTCTCCGTGCAGCCCAACTTCCAGCAGGACAAGTTCCTG
GGGCGCTGGTTCAGCGCGGGCCTCGCCTCCAACTCGAGCTGGCTCCGGGAGAAGAAGGCG
GCGTTGTCCATGTGCAAGTCTGTGGTGGCCCCTGCCACGGATGGTGGCCTCAACCTGACC
TCCACCTTCCTCAGGAAAAACCAGTGTGAGACCCGAACCATGCTGCTGCAGCCCGCGGGG
TCCCTCGGCTCCTACAGCTACCGGAGTCCCCACTGGGGCAGCACCTACTCCGTGTCAGTG
GTGGAGACCGACTACGACCAGTACGCGCTGCTGTACAGCCAGGGCAGCAAGGGCCCTGGC
GAGGACTTCCGCATGGCCACCCTCTACAGCCGAACCCAGACCCCCAGGGCTGAGTTAAAG
GAGAAATTCACCGCCTTCTGCAAGGCCCAGGGCTTCACAGAGGATACCATTGTCTTCCTG
CCCCAAACCGATAAGTGCATGACGGAACAATAG
|
| Protein Properties |
|---|
| Number of Residues
| 190 |
| Molecular Weight
| 21028.665 |
| Theoretical pI
| 7.796 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Prostaglandin-H2 D-isomerase
MATHHTLWMGLALLGVLGDLQAAPEAQVSVQPNFQQDKFLGRWFSAGLASNSSWLREKKA
ALSMCKSVVAPATDGGLNLTSTFLRKNQCETRTMLLQPAGSLGSYSYRSPHWGSTYSVSV
VETDYDQYALLYSQGSKGPGEDFRMATLYSRTQTPRAELKEKFTAFCKAQGFTEDTIVFL
PQTDKCMTEQ
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| P41222 |
| UniProtKB/Swiss-Prot Entry Name
| PTGDS_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| M61900 |
| GeneCard ID
| PTGDS |
| GenAtlas ID
| PTGDS |
| HGNC ID
| HGNC:9592 |
| References |
|---|
| General References
| Not Available |
