| Identification |
|---|
| HMDB Protein ID
| CDBP01443 |
| Secondary Accession Numbers
| Not Available |
| Name
| Cytochrome P450 1A1 |
| Description
| Not Available |
| Synonyms
|
- CYPIA1
- Cytochrome P450 form 6
- Cytochrome P450-C
- Cytochrome P450-P1
|
| Gene Name
| CYP1A1 |
| Protein Type
| Metal Binding |
| Biological Properties |
|---|
| General Function
| Involved in monooxygenase activity |
| Specific Function
| Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
|
| GO Classification
|
| Biological Process |
| response to antibiotic |
| response to drug |
| dibenzo-p-dioxin catabolic process |
| 9-cis-retinoic acid biosynthetic process |
| digestive tract development |
| response to organic cyclic compound |
| embryo development ending in birth or egg hatching |
| cellular lipid metabolic process |
| flavonoid metabolic process |
| response to iron(III) ion |
| hepatocyte differentiation |
| response to hyperoxia |
| hydrogen peroxide biosynthetic process |
| response to vitamin A |
| insecticide metabolic process |
| response to hypoxia |
| maternal process involved in parturition |
| response to arsenic-containing substance |
| response to herbicide |
| porphyrin-containing compound metabolic process |
| drug metabolic process |
| positive regulation of S phase of mitotic cell cycle |
| response to lipopolysaccharide |
| response to food |
| cell proliferation |
| response to nematode |
| vitamin D metabolic process |
| response to virus |
| response to wounding |
| xenobiotic metabolic process |
| camera-type eye development |
| amine metabolic process |
| coumarin metabolic process |
| Cellular Component |
| endoplasmic reticulum membrane |
| mitochondrion |
| Function |
| transition metal ion binding |
| electron carrier activity |
| iron ion binding |
| monooxygenase activity |
| heme binding |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen |
| oxidoreductase activity |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| Molecular Function |
| electron carrier activity |
| oxygen binding |
| aromatase activity |
| demethylase activity |
| flavonoid 3'-monooxygenase activity |
| oxidoreductase activity, acting on diphenols and related substances as donors |
| vitamin D 24-hydroxylase activity |
| iron ion binding |
| steroid hydroxylase activity |
| heme binding |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Peripheral membrane protein
- Peripheral membrane protein
- Endoplasmic reticulum membrane
- Microsome membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Steroid hormone biosynthesis | Not Available |  | | Metabolism of xenobiotics by cytochrome P450 | Not Available |  | | Chemical carcinogenesis | Not Available |  | | Tryptophan Metabolism |   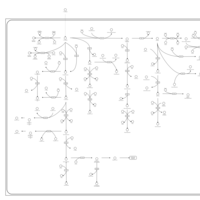 |  | | Estrone Metabolism |   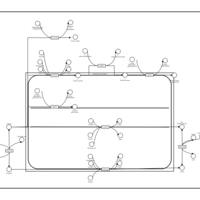 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 15 |
| Locus
| 15q24.1 |
| SNPs
| CYP1A1 |
| Gene Sequence
|
>1539 bp
ATGCTTTTCCCAATCTCCATGTCGGCCACGGAGTTTCTTCTGGCCTCTGTCATCTTCTGT
CTGGTATTCTGGGTAATGAGGGCCTCAAGACCTCAGGTCCCCAAAGGCCTGAAGAATCCA
CCAGGGCCATGGGGCTGGCCTCTGATTGGGCACATGCTGACCCTGGGAAAGAACCCGCAC
CTGGCACTGTCAAGGATGAGCCAGCAGTATGGGGACGTGCTGCAGATCCGAATTGGCTCC
ACACCCGTGGTGGTGCTGAGCGGCCTGGACACCATCCGGCAGGCCCTGGTGCGGCAGGGC
GATGATTTCAAGGGCCGGCCCGACCTCTACACCTTCACCCTCATCAGTAATGGTCAGAGC
ATGTCCTTCAGCCCAGACTCTGGACCAGTGTGGGCTGCCCGCCGGCGCCTGGCCCAGAAT
GGCCTGAAAAGTTTCTCCATTGCCTCTGACCCAGCCTCCTCAACCTCCTGCTACCTGGAA
GAGCATGTGAGCAAGGAGGCTGAGGTCCTGATAAGCACGTTGCAGGAGCTGATGGCAGGG
CCTGGGCACTTTAACCCCTACAGGTATGTGGTGGTATCAGTGACCAATGTCATCTGTGCC
ATTTGCTTTGGCCGGCGCTATGACCACAACCACCAAGAACTGCTTAGCCTAGTCAACCTG
AATAATAATTTCGGGGAGGTGGTTGGCTCTGGAAACCCAGCTGAGTTCATCCCTATTCTT
CGCTACCTACCCAACCCTTCCCTGAATGCCTTCAAGGACCTGAATGAGAAGTTCTACAGC
TTCATGCAGAAGATGGTCAAGGAGCACTACAAAACCTTTGAGAAGGGCCACATCCGGGAC
ATCACAGACAGCCTGATTGAGCACTGTCAGGAGAAGCAGCTGGATGAGAACGCCAATGTC
CAGCTGTCAGATGAGAAGATCATTAACATCGTCTTGGACCTCTTTGGAGCTGGGTTTGAC
ACAGTCACAACTGCTATCTCCTGGAGCCTCATGTATTTGGTGATGAACCCCAGGGTACAG
AGAAAGATCCAAGAGGAGCTAGACACAGTGATTGGCAGGTCACGGCGGCCCCGGCTCTCT
GACAGATCCCATCTGCCCTATATGGAGGCCTTCATCCTGGAGACCTTCCGACACTCTTCC
TTCGTCCCCTTCACCATCCCCCACAGCACAACAAGAGACACAAGTTTGAAAGGCTTTTAC
ATCCCCAAGGGGCGTTGTGTCTTTGTAAACCAGTGGCAGATCAACCATGACCAGAAGCTA
TGGGTCAACCCATCTGAGTTCCTACCTGAACGGTTTCTCACCCCTGATGGTGCTATCGAC
AAGGTGTTAAGTGAGAAGGTGATTATCTTTGGCATGGGCAAGCGGAAGTGTATCGGTGAG
ACCGTTGCCCGCTGGGAGGTCTTTCTCTTCCTGGCTATCCTGCTGCAACGGGTGGAATTC
AGCGTGCCACTGGGCGTGAAGGTGGACATGACCCCCATCTATGGGCTAACCATGAAGCAT
GCCTGCTGTGAGCACTTCCAAATGCAGCTGCGCTCTTAG
|
| Protein Properties |
|---|
| Number of Residues
| 512 |
| Molecular Weight
| 58164.815 |
| Theoretical pI
| 8.347 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Cytochrome P450 1A1
MLFPISMSATEFLLASVIFCLVFWVIRASRPQVPKGLKNPPGPWGWPLIGHMLTLGKNPH
LALSRMSQQYGDVLQIRIGSTPVVVLSGLDTIRQALVRQGDDFKGRPDLYTFTLISNGQS
MSFSPDSGPVWAARRRLAQNGLKSFSIASDPASSTSCYLEEHVSKEAEVLISTLQELMAG
PGHFNPYRYVVVSVTNVICAICFGRRYDHNHQELLSLVNLNNNFGEVVGSGNPADFIPIL
RYLPNPSLNAFKDLNEKFYSFMQKMVKEHYKTFEKGHIRDITDSLIEHCQEKQLDENANV
QLSDEKIINIVLDLFGAGFDTVTTAISWSLMYLVMNPRVQRKIQEELDTVIGRSRRPRLS
DRSHLPYMEAFILETFRHSSFVPFTIPHSTTRDTSLKGFYIPKGRCVFVNQWQINHDQKL
WVNPSEFLPERFLTPDGAIDKVLSEKVIIFGMGKRKCIGETIARWEVFLFLAILLQRVEF
SVPLGVKVDMTPIYGLTMKHACCEHFQMQLRS
|
| External Links |
|---|
| GenBank ID Protein
| 181276 |
| UniProtKB/Swiss-Prot ID
| P04798 |
| UniProtKB/Swiss-Prot Entry Name
| CP1A1_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| K03191 |
| GeneCard ID
| CYP1A1 |
| GenAtlas ID
| CYP1A1 |
| HGNC ID
| HGNC:2595 |
| References |
|---|
| General References
| Not Available |



