| Identification |
|---|
| HMDB Protein ID
| CDBP01010 |
| Secondary Accession Numbers
| Not Available |
| Name
| Retinal guanylyl cyclase 2 |
| Description
| Not Available |
| Synonyms
|
- GC-F
- Guanylate cyclase 2F, retinal
- Guanylate cyclase F
- RETGC-2
- ROS-GC2
- Rod outer segment membrane guanylate cyclase 2
|
| Gene Name
| GUCY2F |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in phosphorus-oxygen lyase activity |
| Specific Function
| Probably plays a specific functional role in the rods and/or cones of photoreceptors. It may be the enzyme involved in the resynthesis of cGMP required for recovery of the dark state after phototransduction.
|
| GO Classification
|
| Biological Process |
| receptor guanylyl cyclase signaling pathway |
| visual perception |
| intracellular signal transduction |
| Cellular Component |
| nuclear outer membrane |
| integral to plasma membrane |
| Function |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| lyase activity |
| cyclase activity |
| phosphorus-oxygen lyase activity |
| protein kinase activity |
| guanylate cyclase activity |
| binding |
| catalytic activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| kinase activity |
| nucleoside binding |
| purine nucleoside binding |
| Molecular Function |
| ATP binding |
| guanylate cyclase activity |
| protein kinase activity |
| GTP binding |
| receptor activity |
| Process |
| purine nucleotide biosynthetic process |
| signaling pathway |
| metabolic process |
| phosphorylation |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| nucleoside monophosphate metabolic process |
| nucleoside monophosphate biosynthetic process |
| cellular metabolic process |
| cyclic nucleotide biosynthetic process |
| protein amino acid phosphorylation |
| cgmp biosynthetic process |
| phosphorus metabolic process |
| intracellular signaling pathway |
| phosphate metabolic process |
| purine nucleotide metabolic process |
| signaling |
|
| Cellular Location
|
- Membrane
- Single-pass type I membrane protein
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Phototransduction | Not Available | 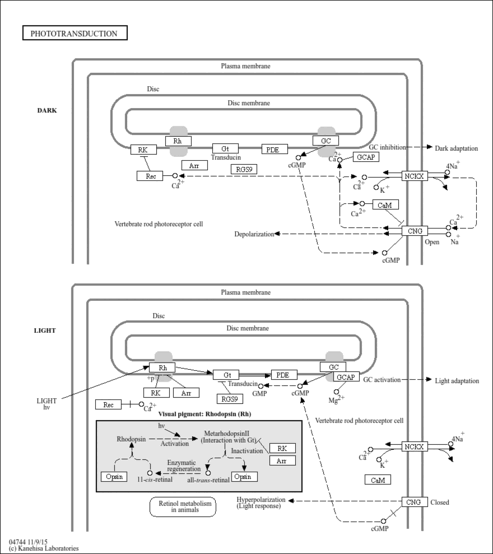 |
|
| Gene Properties |
|---|
| Chromosome Location
| X |
| Locus
| Xq22 |
| SNPs
| GUCY2F |
| Gene Sequence
|
>3327 bp
ATGTTCCTGGGACTCGGGCGCTTTTCTCGCCTTGTTCTCTGGTTTGCGGCTTTCAGGAAA
CTGCTGGGACACCATGGCCTTGCATCTGCCAAGTTCCTGTGGTGCTTGTGCCTTCTGTCT
GTCATGTCCCTTCCGCAGCAGGTGTGGACACTCCCCTACAAGATAGGGGTGGTGGGCCCT
TGGGCTTGTGATTCGCTGTTTTCAAAGGCCCTGCCTGAGGTTGCTGCGCGATTAGCCATT
GAGCGAATCAACCGGGACCCATCTTTTGACCTGAGTTATTCTTTTGAATACGTGATTCTC
AATGAAGACTGCCAGACTTCGAGGGCTCTCTCCAGTTTCATTTCCCACCACCAGATGGCC
TCAGGATTTATTGGACCTACCAACCCTGGCTACTGCGAGGCAGCCTCGCTCCTGGGAAAC
AGCTGGGACAAAGGAATTTTCTCTTGGGCTTGTGTGAATTATGAATTAGACAATAAAATT
AGCTACCCGACCTTTTCTCGGACACTCCCTTCTCCCATCCGGGTGCTTGTAACTGTCATG
AAATATTTCCAGTGGGCTCATGCTGGAGTCATTTCCTCAGATGAAGACATTTGGGTGCAT
ACAGCCAATCGAGTCGCAAGTGCTCTTCGGAGCCACGGCTTACCTGTAGGGGTCGTCCTG
ACCACAGGACAAGACAGCCAAAGCATGCGGAAAGCCCTCCAGAGGATTCACCAGGCAGAC
AGAATTCGCATAATCATCATGTGTATGCATTCAGCTTTGATTGGGGGAGAGACTCAGATG
CATCTCTTGGAATGTGCTCATGATCTGAAAATGACTGATGGAACCTACGTCTTTGTTCCT
TATGATGCCCTGCTCTACAGTTTACCTTATAAGCACACCCCCTACCGGGTCCTAAGGAAC
AACCCAAAGCTCCGGGAAGCCTATGATGCAGTGTTGACCATTACAGTGGAGTCCCAAGAA
AAGACCTTCTATCAAGCCTTCACAGAGGCAGCAGCAAGAGGTGAAATTCCTGAGAAGCTG
GAGTTCGATCAAGTTTCACCGTTGTTTGGAACCATCTACAATTCAATTTACTTTATCGCA
CAAGCCATGAATAATGCTATGAAAGAAAATGGACAGGCTGGTGCTGCCAGCCTGGTTCAG
CATTCCAGAAACATGCAGTTCCATGGATTCAACCAGTTGATGAGGACAGATTCAAATGGA
AATGGAATTTCAGAATATGTAATCCTGGACACCAACTTGAAAGAATGGGAACTCCATAGC
ACCTACACTGTGGACATGGAAATGGAGCTGCTACGTTTCGGAGGGACCCCTATTCACTTC
CCTGGTGGCAGGCCCCCTAGAGCAGATGCAAAATGCTGGTTTGCAGAAGGGAAGATCTGC
CATGGAGGCATCGACCCTGCCTTTGCCATGATGGTCTGCCTTACTTTGCTTATAGCCCTG
CTGTCTATTAATGGATTTGCTTACTTTATAAGGCGTCGTATAAATAAAATCCAGTTGATC
AAAGGACCCAATAGAATTCTACTGACTTTGGAGGATGTAACGTTTATCAATCCCCACTTT
GGCAGTAAGAGAGGAAGTCGTGCCAGTGTAAGCTTCCAGATTACCTCAGAGGTCCAAAGT
GGGAGGTCCCCAAGACTCTCCTTTTCTTCAGGGAGTCTAACTCCAGCTACCTATGAAAAC
TCCAACATAGCGATTTATGAGGGTGATTGGGTGTGGCTGAAAAAGTTCTCCCTTGGAGAT
TTTGGAGACCTTAAGTCCATCAAATCAAGAGCAAGTGATGTGTTCGAAATGATGAAGGAC
TTGCGTCATGAGAATATTAACCCTTTATTGGGTTTCTTCTATGATTCGGGGATGTTTGCC
ATTGTGACAGAATTCTGTTCCCGAGGGAGCCTAGAAGACATACTGACAAATCAAGATGTG
AAACTTGACTGGATGTTTAAATCATCACTCTTGCTGGATCTCATAAAGGGCATGAAGTAC
TTACACCACAGAGAGTTTGTTCATGGGAGGCTAAAGTCTCGAAACTGTGTGGTAGATGGG
CGTTTTGTACTAAAAGTGACAGATTATGGCTTTAACGACATCTTAGAAATGCTGAGACTC
TCTGAAGAGGAATCTTCTATGGAAGAGCTGCTGTGGACGGCCCCTGAACTGTTGAGAGCT
CCAAGAGGCAGCAGGTTAGGTTCTTTTGCAGGAGATGTCTATAGCTTTGCCATCATCATG
CAAGAAGTGATGGTCCGGGGTACCCCATTCTGCATGATGGATCTGCCAGCTCAAGAAATC
ATAAACAGACTTAAGAAGCCTCCTCCTGTGTACAGACCAGTAGTTCCTCCTGAGCATGCC
CCTCCAGAATGTCTCCAGCTGATGAAGCAGTGCTGGGCTGAGGCTGCAGAACAACGACCA
ACTTTTGATGAAATATTTAACCAGTTTAAAACTTTTAATAAAGGGAAGAAGACCAATATT
ATTGATTCTATGCTTCGGATGTTGGAGCAATATTCTAGCAACTTGGAAGATTTGATTCGG
GAGCGGACTGAAGAGCTGGAAATTGAAAAACAGAAAACGGAAAAGCTTCTAACACAGATG
CTACCACCATCAGTTGCTGAATCTCTCAAAAAGGGCTGCACAGTTGAACCTGAGGGCTTT
GACTTGGTCACCTTGTACTTCAGCGACATTGTGGGCTTCACAACCATTTCAGCCATGAGT
GAGCCCATTGAGGTCGTGGATCTTCTGAATGACCTGTACACACTCTTTGATGCAATAATT
GGCAGTCATGATGTCTACAAGGTAGAGACCATTGGAGATGCCTACATGGTGGCTTCAGGC
CTCCCAAAGAGGAATGGCAGTAGGCATGCAGCTGAGATTGCAAACATGTCCTTAGATATC
CTGAGCTCTGTGGGCACTTTCAAGATGCGGCACATGCCAGAAGTGCCGGTCCGAATTCGA
ATTGGCCTTCACTCAGGGCCGGTTGTTGCTGGAGTGGTGGGCCTCACCATGCCCAGATAC
TGCTTGTTTGGAGACACTGTGAACACAGCTTCTCGGATGGAATCTACAGGCTTACCTTAT
CGCATTCATGTCAGTCTCAGCACTGTTACAATTCTTCAAAATCTGAGTGAGGGCTATGAA
GTGGAGCTTCGAGGAAGAACAGAGCTCAAGGGCAAAGGCACAGAGGAAACCTTCTGGCTG
ATTGGGAAAAAAGGCTTCATGAAGCCCCTTCCTGTGCCCCCACCAGTGGACAAAGATGGG
CAAGTGGGCCATGGCCTGCAACCAGTGGAGATTGCAGCCTTCCAAAGAAGAAAAGCAGAA
AGGCAGTTGGTGAGAAACAAGCCATAA
|
| Protein Properties |
|---|
| Number of Residues
| 1108 |
| Molecular Weight
| 124848.965 |
| Theoretical pI
| 7.274 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Retinal guanylyl cyclase 2
MFLGLGRFSRLVLWFAAFRKLLGHHGLASAKFLWCLCLLSVMSLPQQVWTLPYKIGVVGP
WACDSLFSKALPEVAARLAIERINRDPSFDLSYSFEYVILNEDCQTSRALSSFISHHQMA
SGFIGPTNPGYCEAASLLGNSWDKGIFSWACVNYELDNKISYPTFSRTLPSPIRVLVTVM
KYFQWAHAGVISSDEDIWVHTANRVASALRSHGLPVGVVLTTGQDSQSMRKALQRIHQAD
RIRIIIMCMHSALIGGETQMHLLECAHDLKMTDGTYVFVPYDALLYSLPYKHTPYQVLRN
NPKLREAYDAVLTITVESQEKTFYQAFTEAAARGEIPEKLEFDQVSPLFGTIYNSIYFIA
QAMNNAMKENGQAGAASLVQHSRNMQFHGFNQLMRTDSNGNGISEYVILDTNLKEWELHS
TYTVDMEMELLRFGGTPIHFPGGRPPRADAKCWFAEGKICHGGIDPAFAMMVCLTLLIAL
LSINGFAYFIRRRINKIQLIKGPNRILLTLEDVTFINPHFGSKRGSRASVSFQITSEVQS
GRSPRLSFSSGSLTPATYENSNIAIYEGDWVWLKKFSLGDFGDLKSIKSRASDVFEMMKD
LRHENINPLLGFFYDSGMFAIVTEFCSRGSLEDILTNQDVKLDWMFKSSLLLDLIKGMKY
LHHREFVHGRLKSRNCVVDGRFVLKVTDYGFNDILEMLRLSEEESSMEELLWTAPELLRA
PRGSRLGSFAGDVYSFAIIMQEVMVRGTPFCMMDLPAQEIINRLKKPPPVYRPVVPPEHA
PPECLQLMKQCWAEAAEQRPTFDEIFNQFKTFNKGKKTNIIDSMLRMLEQYSSNLEDLIR
ERTEELEIEKQKTEKLLTQMLPPSVAESLKKGCTVEPEGFDLVTLYFSDIVGFTTISAMS
EPIEVVDLLNDLYTLFDAIIGSHDVYKVETIGDAYMVASGLPKRNGSRHAAEIANMSLDI
LSSVGTFKMRHMPEVPVRIRIGLHSGPVVAGVVGLTMPRYCLFGDTVNTASRMESTGLPY
RIHVSLSTVTILQNLSEGYEVELRGRTELKGKGTEETFWLIGKKGFMKPLPVPPPVDKDG
QVGHGLQPVEIAAFQRRKAERQLVRNKP
|
| External Links |
|---|
| GenBank ID Protein
| 4680397 |
| UniProtKB/Swiss-Prot ID
| P51841 |
| UniProtKB/Swiss-Prot Entry Name
| GUC2F_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AL031387 |
| GeneCard ID
| GUCY2F |
| GenAtlas ID
| GUCY2F |
| HGNC ID
| HGNC:4691 |
| References |
|---|
| General References
| Not Available |
