| Identification |
|---|
| HMDB Protein ID
| CDBP00907 |
| Secondary Accession Numbers
| Not Available |
| Name
| DNA polymerase iota |
| Description
| Not Available |
| Synonyms
|
- Eta2
- RAD30 homolog B
|
| Gene Name
| POLI |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in damaged DNA binding |
| Specific Function
| Error-prone DNA polymerase specifically involved in DNA repair. Plays an important role in translesion synthesis, where the normal high-fidelity DNA polymerases cannot proceed and DNA synthesis stalls. Favors Hoogsteen base-pairing in the active site. Inserts the correct base with high-fidelity opposite an adenosine template. Exhibits low fidelity and efficiency opposite a thymidine template, where it will preferentially insert guanosine. May play a role in hypermutation of immunogobulin genes. Forms a Schiff base with 5'-deoxyribose phosphate at abasic sites, but may not have lyase activity.
|
| GO Classification
|
| Biological Process |
| DNA repair |
| DNA replication |
| Cellular Component |
| nucleoplasm |
| Function |
| binding |
| catalytic activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| nucleotidyltransferase activity |
| dna polymerase activity |
| dna-directed dna polymerase activity |
| damaged dna binding |
| nucleic acid binding |
| dna binding |
| Molecular Function |
| damaged DNA binding |
| metal ion binding |
| DNA-directed DNA polymerase activity |
| Process |
| metabolic process |
| macromolecule metabolic process |
| cellular macromolecule metabolic process |
| dna metabolic process |
| dna repair |
|
| Cellular Location
|
- Nucleus
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Fanconi anemia pathway | Not Available | 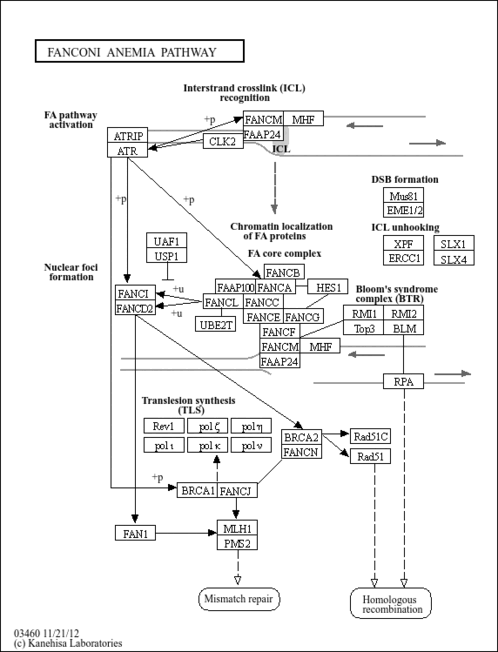 |
|
| Gene Properties |
|---|
| Chromosome Location
| 18 |
| Locus
| 18q21.1 |
| SNPs
| POLI |
| Gene Sequence
|
>2148 bp
ATGGAACTGGCGGACGTGGGGGCGGCAGCCAGCTCGCAGGGAGTTCATGATCAAGTGTTG
CCCACACCAAATGCTTCATCCAGAGTCATAGTACATGTGGATCTGGATTGCTTTTATGCA
CAAGTAGAAATGATCTCAAATCCAGAGCTAAAAGACAAACCTTTAGGGGTTCAACAGAAA
TATTTGGTGGTTACCTGCAACTATGAAGCTAGGAAACTTGGAGTTAAGAAACTTATGAAT
GTCAGAGATGCAAAAGAAAAGTGTCCACAGTTGGTATTAGTTAATGGAGAAGACCTGACC
CGCTACAGAGAAATGTCTTATAAGGTTACAGAATTACTGGAAGAATTTAGTCCAGTTGTT
GAGAGACTTGGATTTGATGAAAATTTTGTGGATCTAACAGAAATGGTTGAGAAGAGACTA
CAGCAGCTGCAAAGTGATGAACTTTCTGCGGTGACTGTGTCGGGTCATGTATACAATAAT
CAGTCTATAAACCTGCTTGACGTCTTGCACATCAGACTACTTGTTGGATCTCAGATTGCA
GCAGAGATGCGGGAAGCCATGTATAATCAGTTGGGGCTCACTGGCTGTGCTGGAGTGGCT
TCTAATAAACTGTTGGCAAAATTAGTTTCTGGTGTCTTTAAACCAAATCAACAAACAGTC
TTATTACCTGAAAGTTGTCAACATCTTATTCATAGTTTGAATCACATAAAGGAAATACCT
GGTATTGGCTATAAAACTGCCAAATGTCTTGAAGCACTGGGTATCAATAGTGTGCGTGAT
CTCCAAACCTTTTCACCCAAAATTTTAGAAAAAGAATTAGGAATTTCAGTTGCTCAGCGT
ATCCAAAAGCTCAGTTTTGGAGAGGATAACTCCCCTGTGATACTCTCAGGACCACCTCAG
TCCTTTAGTGAAGAAGATTCATTTAAAAAATGTTCATCTGAAGTTGAAGCTAAAAATAAG
ATTGAAGAACTACTTGCTAGTCTTTTAAACAGAGTATGCCAAGATGGAAGGAAGCCTCAT
ACAGTGAGATTAATAATCCGTCGGTATTCCTCTGAGAAGCACTATGGTCGTGAGAGTCGT
CAGTGCCCTATTCCTTCACATGTAATTCAGAAATTAGGGACAGGAAATTATGATGTGATG
ACCCCAATGGTTGATATACTTATGAAACTTTTTCGAAATATGGTGAATGTGAAGATGCCA
TTTCACCTTACCCTTCTAAGTGTGTGCTTCTGCAACCTTAAAGCACTAAATACTGCTAAG
AAAGGGCTTATTGATTATTATTTAATGCCATCATTATCAACTACTTCACGCTCTGGCAAG
CACAGTTTTAAAATGAAAGACACTCATATGGAAGATTTTCCCAAAGACAAAGAAACAAAC
CGGGATTTCCTACCAAGTGGAAGAATTGAAAGTACAAGAACTAGGGAGTCTCCACTAGAT
ACCACAAATTTTTCTAAAGAAAAAGACATTAATGAATTCCCACTCTGTTCACTTCCTGAA
GGTGTTGACCAAGAAGTCTTCAAGCAGCTTCCAGTAGATATTCAAGAAGAAATCCTTTCT
GGAAAATCTAGGGAAAAATTTCAAGGGAAAGGAAGTGTGAGTTGTCCATTACATGCCTCT
AGAGGAGTATTATCTTTCTTTTCTAAAAAACAAATGCAAGATATTCCCATAAATCCTAGA
GATCATTTATCCAGTAGCAAACAGGTATCCTCTGTATCTCCTTGTGAACCGGGAACATCA
GGCTTTAATAGCAGTAGTTCTTCTTACATGTCTAGCCAAAAGGATTATTCATATTATTTA
GATAATAGATTAAAAGATGAACGAATAAGTCAAGGACCTAAAGAACCTCAAGGATTCCAC
TTTACAAATTCAAACCCTGCTGTGTCTGCTTTTCATTCATTTCCAAACTTGCAGAGTGAG
CAACTTTTCTCCAGAAACCACACTACAGATAGCCATAAGCAAACAGTAGCAACAGACTCT
CATGAAGGACTTACAGAAAATAGAGAGCCAGATTCTGTTGATGAGAAAATTACTTTCCCT
TCTGACATTGATCCTCAAGTTTTCTATGAACTACCAGAAGCAGTACAAAAGGAACTGCTG
GCAGAGTGGAAGAGAACAGGATCAGATTTCCACATTGGACATAAATAA
|
| Protein Properties |
|---|
| Number of Residues
| 715 |
| Molecular Weight
| 83005.335 |
| Theoretical pI
| 6.08 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>DNA polymerase iota
MELADVGAAASSQGVHDQVLPTPNASSRVIVHVDLDCFYAQVEMISNPELKDKPLGVQQK
YLVVTCNYEARKLGVKKLMNVRDAKEKCPQLVLVNGEDLTRYREMSYKVTELLEEFSPVV
ERLGFDENFVDLTEMVEKRLQQLQSDELSAVTVSGHVYNNQSINLLDVLHIRLLVGSQIA
AEMREAMYNQLGLTGCAGVASNKLLAKLVSGVFKPNQQTVLLPESCQHLIHSLNHIKEIP
GIGYKTAKCLEALGINSVRDLQTFSPKILEKELGISVAQRIQKLSFGEDNSPVILSGPPQ
SFSEEDSFKKCSSEVEAKNKIEELLASLLNRVCQDGRKPHTVRLIIRRYSSEKHYGRESR
QCPIPSHVIQKLGTGNYDVMTPMVDILMKLFRNMVNVKMPFHLTLLSVCFCNLKALNTAK
KGLIDYYLMPSLSTTSRSGKHSFKMKDTHMEDFPKDKETNRDFLPSGRIESTRTRESPLD
TTNFSKEKDINEFPLCSLPEGVDQEVFKQLPVDIQEEILSGKSREKFQGKGSVSCPLHAS
RGVLSFFSKKQMQDIPINPRDHLSSSKQVSSVSPCEPGTSGFNSSSSSYMSSQKDYSYYL
DNRLKDERISQGPKEPQGFHFTNSNPAVSAFHSFPNLQSEQLFSRNHTTDSHKQTVATDS
HEGLTENREPDSVDEKITFPSDIDPQVFYELPEAVQKELLAEWKRTGSDFHIGHK
|
| External Links |
|---|
| GenBank ID Protein
| 5739208 |
| UniProtKB/Swiss-Prot ID
| Q9UNA4 |
| UniProtKB/Swiss-Prot Entry Name
| POLI_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF140501 |
| GeneCard ID
| POLI |
| GenAtlas ID
| POLI |
| HGNC ID
| HGNC:9182 |
| References |
|---|
| General References
| Not Available |
