| Identification |
|---|
| HMDB Protein ID
| CDBP00738 |
| Secondary Accession Numbers
| Not Available |
| Name
| Sarcosine dehydrogenase, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- BPR-2
- SarDH
|
| Gene Name
| SARDH |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Not Available |
| GO Classification
|
| Biological Process |
| glycine catabolic process |
| Cellular Component |
| mitochondrial matrix |
| mitochondrion |
| Component |
| cell part |
| intracellular part |
| cytoplasm |
| Function |
| catalytic activity |
| transferase activity |
| aminomethyltransferase activity |
| oxidoreductase activity |
| transferase activity, transferring one-carbon groups |
| methyltransferase activity |
| Molecular Function |
| sarcosine dehydrogenase activity |
| aminomethyltransferase activity |
| flavin adenine dinucleotide binding |
| Process |
| metabolic process |
| cellular metabolic process |
| serine family amino acid metabolic process |
| glycine metabolic process |
| glycine catabolic process |
| cellular amino acid and derivative metabolic process |
| cellular amino acid metabolic process |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | sarcosine degradation | Not Available | Not Available | | Glycine, serine and threonine metabolism | Not Available |  | | Glycine and Serine Metabolism | 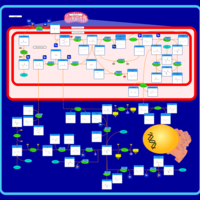 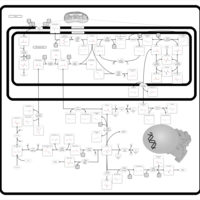 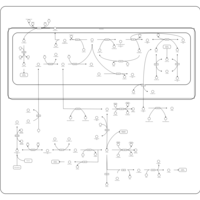 |  | | Dimethylglycine Dehydrogenase Deficiency | 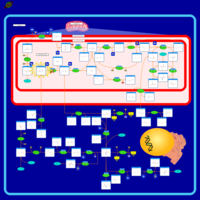 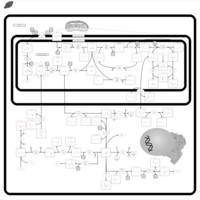  | Not Available | | Dihydropyrimidine Dehydrogenase Deficiency (DHPD) | 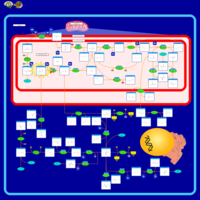 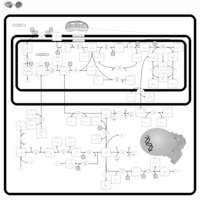 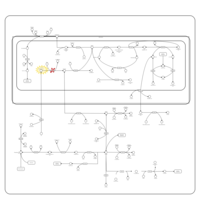 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 9 |
| Locus
| 9q33-q34 |
| SNPs
| SARDH |
| Gene Sequence
|
>2757 bp
ATGGCCTCACTGAGCCGAGCCCTACGTGTGGCTGCTGCCCACCCTCGCCAGAGCCCTACC
CGGGGCATGGGGCCATGCAACCTGTCCAGCGCAGCTGGCCCCACAGCCGAGAAGAGTGTG
CCATATCAGCGGACCCTGAAGGAGGGACAGGGCACCTCGGTGGTGGCCCAAGGCCCAAGC
CGGCCCCTGCCCAGCACGGCCAACGTGGTGGTCATTGGTGGAGGCAGCTTGGGCTGCCAG
ACCCTGTACCACCTGGCCAAGCTGGGCATGAGTGGGGCGGTGCTGCTGGAGCGGGAGCGG
CTGACCTCCGGGACCACCTGGCACACGGCAGGCCTGCTGTGGCAGCTGCGGCCCAGTGAC
GTGGAGGTGGAGCTTCTGGCCCACACTCGGCGGGTGGTGAGCCGGGAGCTGGAGGAGGAG
ACGGGACTACACACGGGCTGGATCCAGAATGGGGGCCTCTTCATCGCGTCCAACCGGCAG
CGCCTGGACGAGTACAAGAGGCTCATGTCGCTGGGCAAGGCGTATGGTGTGGAATCCCAT
GTGCTGAGCCCGGCAGAGACCAAGACTCTGTACCCGCTGATGAATGTGGACGACCTCTAC
GGGACCCTGTATGTGCCGCACGACGGTACCATGGACCCCGCTGGCACCTGTACCACCCTC
GCCAGGGCAGCTTCTGCCCGAGGAGCACAGGTCATTGAGAACTGCCCAGTGACCGGCATT
CGTGTGTGGACGGATGATTTTGGGGTGCGGCGGGTCGCGGGTGTGGAGACTCAGCATGGT
TCCATCCAGACACCCTGCGTGGTCAACTGTGCAGGAGTGTGGGCAAGTGCTGTGGGCCGG
ATGGCTGGAGTCAAGGTCCCGCTGGTGGCCATGCACCATGCCTATGTCGTCACCGAGCGC
ATCGAGGGGATTCAGAACATGCCCAATGTCCGTGATCATGATGCCTCTGTCTACCTCCGC
CTCCAAGGGGATGCCTTGTCTGTGGGTGGCTATGAGGCCAACCCCATCTTTTGGGAGGAG
GTGTCAGACAAGTTTGCCTTCGGCCTCTTTGACCTGGACTGGGAGGTGTTCACCCAGCAC
ATTGAAGGCGCCATCAACAGGGTCCCCGTGCTGGAGAAGACAGGAATCAAGTCCACGGTC
TGCGGCCCTGAATCCTTCACGCCCGACCACAAGCCCCTGATGGGGGAGGCACCTGAGCTC
CGAGGGTTCTTCCTGGGCTGTGGCTTCAACAGCGCAGGAATGATGCTGGGTGGTGGCTGT
GGGCAGGAGCTGGCCCACTGGATCATCCATGGGCGCCCGGAGAAGGACATGCATGGCTAT
GACATCAGGCGCTTCCATCACTCGCTCACGGACCACCCCCGCTGGATCCGAGAGCGAAGC
CATGAGTCCTACGCCAAGAACTACTCCGTCGTCTTCCCCCACGATGAGCCGCTGGCCGGG
CGCAACATGAGGAGAGACCCGCTGCATGAGGAACTCCTTGGACAAGGCTGCGTGTTCCAG
GAGCGGCATGGCTGGGAGCGACCGGGATGGTTTCATCCCCGAGGCCCAGCTCCGGTCCTC
GAGTACGACTACTACGGGGCTTACGGGAGCCGCGCGCACGAGGACTACGCCTACCGCAGG
CTGCTGGCAGACGAGTACACCTTCGCCTTCCCGCCCCACCACGACACGATCAAGAAGGAG
TGCCTGGCCTGCAGAGGGGCCGCCGCTGTGTTTGACATGTCCTACTTCGGGAAGTTCTAC
CTGGTGGGGCTGGATGCAAGGAAGGCTGCCGACTGGCTCTTCTCCGCAGATGTCAGCCGA
CCCCCAGGCTCCACCGTGTACACGTGCATGCTCAACCACCGTGGGGGCACCGAGAGTGAC
CTGACTGTCAGCCGCCTGGCACCCAGCCACCAGGCCTCCCCGCTGGCCCCCGCCTTTGAA
GGGGACGGTTACTACCTGGCCATGGGCGGGGCCGTGGCCCAGCACAACTGGTCCCACATC
ACCACCGTGCTGCAGGACCAGAAGTCCCAGTGCCAGCTCATCGACAGCTCCGAGGACCTG
GGTATGATCAGTATCCAGGGCCCAGCCAGCCGAGCCATTTTGCAGGAGGTGCTGGACGCA
GACCTGAGCAACGAGGCCTTCCCGTTCTCCACCCACAAGCTACTGAGAGCCGCAGGGCAC
CTGGTCCGAGCCATGCGGCTGTCCTTTGTGGGGGAGCTGGGCTGGGAGCTGCACATTCCA
AAGGCGTCCTGCGTGCCTGTGTACCGGGCTGTGATGGCCGCGGGTGCCAAGCACGGCCTC
ATCAACGCAGGGTACCGCGCCATCGACTCCCTGAGCATTGAGAAAGGCTACCGGCACTGG
CACGCGGACCTGCGGCCAGACGACAGCCCCCTGGAGGCAGGCCTGGCCTTCACCTGCAAG
CTCAAGTCGCCGGTGCCCTTCCTGGGGAGGGAGGCCCTGGAGCAGCAGCGGGCCGCAGGC
CTCCGCCGGCGCCTGGTGTGCTTCACCATGGAGGACAAAGTACCCATGTTTGGCCTGGAG
GCCATCTGGAGGAACGGCCAAGTGGTGGGCCATGTCCGGAGGGCTGACTTTGGGTTCGCC
ATCGACAAGACCATCGCCTACGGTTACATCCATGACCCCAGCGGTGGGCCGGTCTCGCTG
GACTTTGTGAAGAGCGGGGACTATGCCCTGGAGAGAATGGGGGTGACCTATGGTGCCCAG
GCTCACCTGAAGTCGCCCTTCGACCCCAACAACAAGAGGGTGAAGGGAATCTACTGA
|
| Protein Properties |
|---|
| Number of Residues
| 918 |
| Molecular Weight
| 101035.985 |
| Theoretical pI
| 7.251 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Sarcosine dehydrogenase, mitochondrial
MASLSRALRVAAAHPRQSPTRGMGPCNLSSAAGPTAEKSVPYQRTLKEGQGTSVVAQGPS
RPLPSTANVVVIGGGSLGCQTLYHLAKLGMSGAVLLERERLTSGTTWHTAGLLWQLRPSD
VEVELLAHTRRVVSRELEEETGLHTGWIQNGGLFIASNRQRLDEYKRLMSLGKAYGVESH
VLSPAETKTLYPLMNVDDLYGTLYVPHDGTMDPAGTCTTLARAASARGAQVIENCPVTGI
RVWTDDFGVRRVAGVETQHGSIQTPCVVNCAGVWASAVGRMAGVKVPLVAMHHAYVVTER
IEGIQNMPNVRDHDASVYLRLQGDALSVGGYEANPIFWEEVSDKFAFGLFDLDWEVFTQH
IEGAINRVPVLEKTGIKSTVCGPESFTPDHKPLMGEAPELRGFFLGCGFNSAGMMLGGGC
GQELAHWIIHGRPEKDMHGYDIRRFHHSLTDHPRWIRERSHESYAKNYSVVFPHDEPLAG
RNMRRDPLHEELLGQGCVFQERHGWERPGWFHPRGPAPVLEYDYYGAYGSRAHEDYAYRR
LLADEYTFAFPPHHDTIKKECLACRGAAAVFDMSYFGKFYLVGLDARKAADWLFSADVSR
PPGSTVYTCMLNHRGGTESDLTVSRLAPSHQASPLAPAFEGDGYYLAMGGAVAQHNWSHI
TTVLQDQKSQCQLIDSSEDLGMISIQGPASRAILQEVLDADLSNEAFPFSTHKLLRAAGH
LVRAMRLSFVGELGWELHIPKASCVPVYRAVMAAGAKHGLINAGYRAIDSLSIEKGYRHW
HADLRPDDSPLEAGLAFTCKLKSPVPFLGREALEQQRAAGLRRRLVCFTMEDKVPMFGLE
AIWRNGQVVGHVRRADFGFAIDKTIAYGYIHDPSGGPVSLDFVKSGDYALERMGVTYGAQ
AHLKSPFDPNNKRVKGIY
|
| External Links |
|---|
| GenBank ID Protein
| 5902974 |
| UniProtKB/Swiss-Prot ID
| Q9UL12 |
| UniProtKB/Swiss-Prot Entry Name
| SARDH_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AF095735 |
| GeneCard ID
| SARDH |
| GenAtlas ID
| SARDH |
| HGNC ID
| HGNC:10536 |
| References |
|---|
| General References
| Not Available |
