| Identification |
|---|
| HMDB Protein ID
| CDBP00620 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ubiquitin-conjugating enzyme E2 N |
| Description
| Not Available |
| Synonyms
|
- Bendless-like ubiquitin-conjugating enzyme
- Ubc13
- Ubiquitin carrier protein N
- Ubiquitin-protein ligase N
- UbcH13
|
| Gene Name
| UBE2N |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acid-amino acid ligase activity |
| Specific Function
| The UBE2V1-UBE2N and UBE2V2-UBE2N heterodimers catalyze the synthesis of non-canonical 'Lys-63'-linked polyubiquitin chains. This type of polyubiquitination does not lead to protein degradation by the proteasome. Mediates transcriptional activation of target genes. Plays a role in the control of progress through the cell cycle and differentiation. Plays a role in the error-free DNA repair pathway and contributes to the survival of cells after DNA damage. Acts together with the E3 ligases, HLTF and SHPRH, in the 'Lys-63'-linked poly-ubiquitination of PCNA upon genotoxic stress, which is required for DNA repair. Appears to act together with E3 ligase RNF5 in the 'Lys-63'-linked polyubiquitination of JKAMP thereby regulating JKAMP function by decreasing its association with components of the proteasome and ERAD. Promotes TRIM5 capsid-specific restriction activity and the UBE2V1-UBE2N heterodimer acts in concert with TRIM5 to generate 'Lys-63'-linked polyubiquitin chains which activate the MAP3K7/TAK1 complex which in turn results in the induction and expression of NF-kappa-B and MAPK-responsive inflammatory genes (By similarity).
|
| GO Classification
|
| Biological Process |
| Toll signaling pathway |
| protein K63-linked ubiquitination |
| toll-like receptor 1 signaling pathway |
| DNA double-strand break processing |
| toll-like receptor 2 signaling pathway |
| double-strand break repair via homologous recombination |
| T cell receptor signaling pathway |
| positive regulation of NF-kappaB transcription factor activity |
| histone ubiquitination |
| toll-like receptor 4 signaling pathway |
| nucleotide-binding oligomerization domain containing signaling pathway |
| positive regulation of histone modification |
| positive regulation of ubiquitin-protein ligase activity |
| regulation of histone ubiquitination |
| innate immune response |
| proteolysis |
| positive regulation of I-kappaB kinase/NF-kappaB cascade |
| positive regulation of DNA repair |
| cytokine-mediated signaling pathway |
| postreplication repair |
| MyD88-dependent toll-like receptor signaling pathway |
| Cellular Component |
| cytosol |
| nucleus |
| ubiquitin ligase complex |
| UBC13-MMS2 complex |
| UBC13-UEV1A complex |
| Function |
| catalytic activity |
| small conjugating protein ligase activity |
| ligase activity |
| ligase activity, forming carbon-nitrogen bonds |
| acid-amino acid ligase activity |
| Molecular Function |
| ubiquitin-protein ligase activity |
| ATP binding |
| ubiquitin binding |
| Process |
| metabolic process |
| regulation of protein metabolic process |
| macromolecule metabolic process |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| post-translational protein modification |
| macromolecule modification |
| protein modification process |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protein ubiquitination | Not Available | Not Available | | Ubiquitin mediated proteolysis | Not Available | 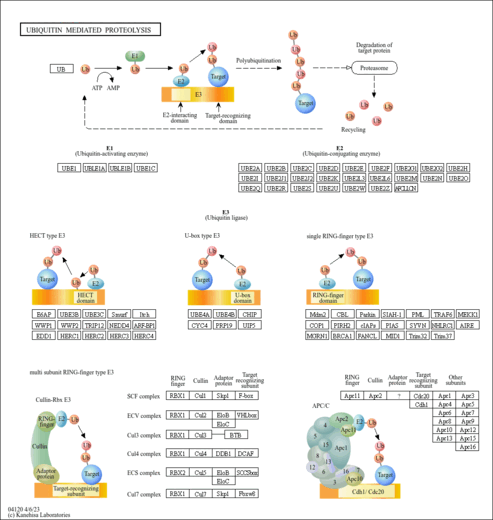 |
|
| Gene Properties |
|---|
| Chromosome Location
| 12 |
| Locus
| 12q22 |
| SNPs
| UBE2N |
| Gene Sequence
|
>459 bp
ATGGCCGGGCTGCCCCGCAGGATCATCAAGGAAACCCAGCGTTTGCTGGCAGAACCAGTT
CCTGGCATCAAAGCCGAACCAGATGAGAGCAACGCCCGTTATTTTCATGTGGTCATTGCT
GGCCCTCAGGATTCCCCCTTTGAGGGAGGGACTTTTAAACTTGAACTATTCCTTCCAGAA
GAATACCCAATGGCAGCCCCTAAAGTACGTTTCATGACCAAAATTTATCATCCTAATGTA
GACAAGTTGGGAAGAATATGTTTAGATATTTTGAAAGATAAGTGGTCCCCAGCACTGCAG
ATCCGCACAGTTCTGCTATCGATCCAGGCCTTGTTAAGTGCTCCCAATCCAGATGATCCA
TTAGCAAATGATGTAGCGGAGCAGTGGAAGACCAACGAAGCCCAAGCCATAGAAACAGCT
AGAGCATGGACTAGGCTATATGCCATGAATAATATTTAA
|
| Protein Properties |
|---|
| Number of Residues
| 152 |
| Molecular Weight
| 17137.625 |
| Theoretical pI
| 6.567 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ubiquitin-conjugating enzyme E2 N
MAGLPRRIIKETQRLLAEPVPGIKAEPDESNARYFHVVIAGPQDSPFEGGTFKLELFLPE
EYPMAAPKVRFMTKIYHPNVDKLGRICLDILKDKWSPALQIRTVLLSIQALLSAPNPDDP
LANDVAEQWKTNEAQAIETARAWTRLYAMNNI
|
| External Links |
|---|
| GenBank ID Protein
| 12653255 |
| UniProtKB/Swiss-Prot ID
| P61088 |
| UniProtKB/Swiss-Prot Entry Name
| UBE2N_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| BC000396 |
| GeneCard ID
| UBE2N |
| GenAtlas ID
| UBE2N |
| HGNC ID
| HGNC:12492 |
| References |
|---|
| General References
| Not Available |
