| Identification |
|---|
| HMDB Protein ID
| CDBP00602 |
| Secondary Accession Numbers
| Not Available |
| Name
| SUMO-conjugating enzyme UBC9 |
| Description
| Not Available |
| Synonyms
|
- SUMO-protein ligase
- Ubiquitin carrier protein 9
- Ubiquitin carrier protein I
- Ubiquitin-conjugating enzyme E2 I
- Ubiquitin-protein ligase I
- p18
|
| Gene Name
| UBE2I |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acid-amino acid ligase activity |
| Specific Function
| Accepts the ubiquitin-like proteins SUMO1, SUMO2, SUMO3 and SUMO4 from the UBLE1A-UBLE1B E1 complex and catalyzes their covalent attachment to other proteins with the help of an E3 ligase such as RANBP2 or CBX4. Can catalyze the formation of poly-SUMO chains. Necessary for sumoylation of FOXL2 and KAT5. Essential for nuclear architecture and chromosome segregation.
|
| GO Classification
|
| Biological Process |
| positive regulation of intracellular steroid hormone receptor signaling pathway |
| positive regulation of sequence-specific DNA binding transcription factor activity |
| virus-host interaction |
| negative regulation of transcription from RNA polymerase II promoter |
| protein sumoylation |
| regulation of receptor activity |
| proteasomal ubiquitin-dependent protein catabolic process |
| mitosis |
| cell division |
| ubiquitin-dependent protein catabolic process |
| chromosome segregation |
| Cellular Component |
| cytoplasm |
| dendrite |
| fibrillar center |
| synaptonemal complex |
| PML body |
| synapse |
| Function |
| ligase activity |
| ligase activity, forming carbon-nitrogen bonds |
| acid-amino acid ligase activity |
| catalytic activity |
| small conjugating protein ligase activity |
| Molecular Function |
| ubiquitin-protein ligase activity |
| ATP binding |
| SUMO ligase activity |
| Process |
| metabolic process |
| regulation of protein metabolic process |
| macromolecule metabolic process |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| post-translational protein modification |
| macromolecule modification |
| protein modification process |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protein sumoylation | Not Available | Not Available | | RNA transport | Not Available | 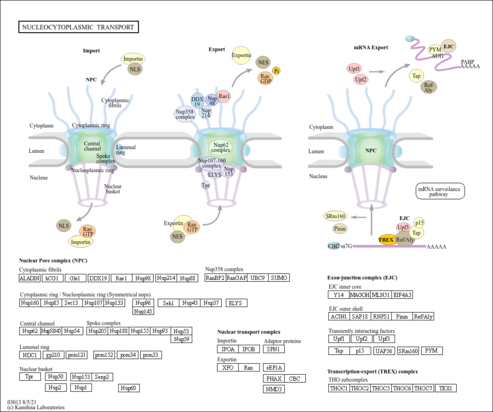 | | NF-kappa B signaling pathway | Not Available | 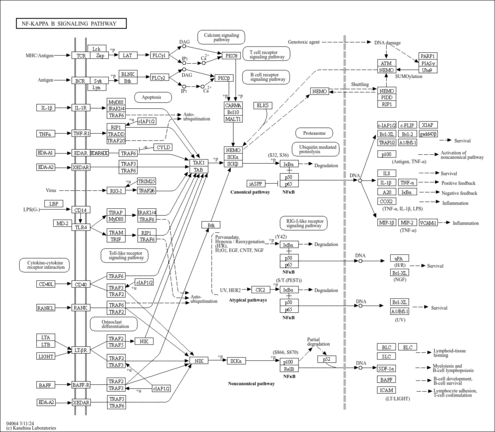 | | Ubiquitin mediated proteolysis | Not Available | 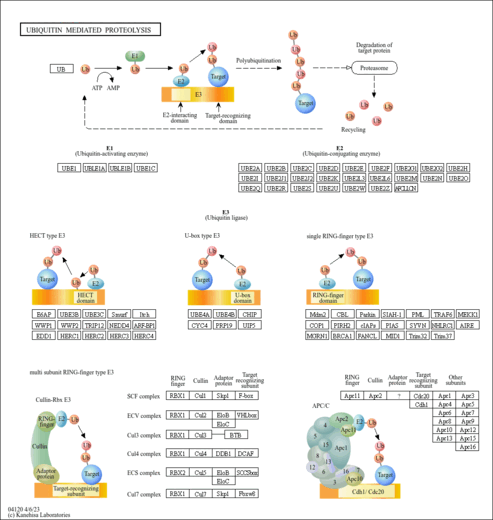 |
|
| Gene Properties |
|---|
| Chromosome Location
| 16 |
| Locus
| 16p13.3 |
| SNPs
| UBE2I |
| Gene Sequence
|
>477 bp
ATGTCGGGGATCGCCCTCAGCAGACTCGCCCAGGAGAGGAAAGCATGGAGGAAAGACCAC
CCATTTGGTTTCGTGGCTGTCCCAACAAAAAATCCCGATGGCACGATGAACCTCATGAAC
TGGGAGTGCGCCATTCCAGGAAAGAAAGGGACTCCGTGGGAAGGAGGCTTGTTTAAACTA
CGGATGCTTTTCAAAGATGATTATCCATCTTCGCCACCAAAATGTAAATTCGAACCACCA
TTATTTCACCCGAATGTGTACCCTTCGGGGACAGTGTGCCTGTCCATCTTAGAGGAGGAC
AAGGACTGGAGGCCAGCCATCACAATCAAACAGATCCTATTAGGAATACAGGAACTTCTA
AATGAACCAAATATCCAAGACCCAGCTCAAGCAGAGGCCTACACGATTTACTGCCAAAAC
AGAGTGGAGTACGAGAAAAGGGTCCGAGCACAAGCCAAGAAGTTTGCGCCCTCATAA
|
| Protein Properties |
|---|
| Number of Residues
| 158 |
| Molecular Weight
| 18006.665 |
| Theoretical pI
| 8.659 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>SUMO-conjugating enzyme UBC9
MSGIALSRLAQERKAWRKDHPFGFVAVPTKNPDGTMNLMNWECAIPGKKGTPWEGGLFKL
RMLFKDDYPSSPPKCKFEPPLFHPNVYPSGTVCLSILEEDKDWRPAITIKQILLGIQELL
NEPNIQDPAQAEAYTIYCQNRVEYEKRVRAQAKKFAPS
|
| External Links |
|---|
| GenBank ID Protein
| 4507785 |
| UniProtKB/Swiss-Prot ID
| P63279 |
| UniProtKB/Swiss-Prot Entry Name
| UBC9_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_003345.3 |
| GeneCard ID
| UBE2I |
| GenAtlas ID
| UBE2I |
| HGNC ID
| HGNC:12485 |
| References |
|---|
| General References
| Not Available |


