| Identification |
|---|
| HMDB Protein ID
| CDBP00568 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ubiquitin-conjugating enzyme E2 L3 |
| Description
| Not Available |
| Synonyms
|
- L-UBC
- UbcH7
- Ubiquitin carrier protein L3
- Ubiquitin-conjugating enzyme E2-F1
- Ubiquitin-protein ligase L3
|
| Gene Name
| UBE2L3 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acid-amino acid ligase activity |
| Specific Function
| Ubiquitin-conjugating enzyme E2 that specifically acts with HECT-type and RBR family E3 ubiquitin-protein ligases. Does not function with most RING-containing E3 ubiquitin-protein ligases because it lacks intrinsic E3-independent reactivity with lysine: in contrast, it has activity with the RBR family E3 enzymes, such as PARK2 and ARIH1, that function like function like RING-HECT hybrids. Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. In vitro catalyzes 'Lys-11'-linked polyubiquitination. Involved in the selective degradation of short-lived and abnormal proteins. Down-regulated during the S-phase it is involved in progression through the cell cycle. Regulates nuclear hormone receptors transcriptional activity. May play a role in myelopoiesis.
|
| GO Classification
|
| Biological Process |
| regulation of S phase of mitotic cell cycle |
| cellular response to glucocorticoid stimulus |
| regulation of transcription, DNA-dependent |
| transcription, DNA-dependent |
| cell proliferation |
| ubiquitin-dependent protein catabolic process |
| protein K11-linked ubiquitination |
| Cellular Component |
| cytoplasm |
| nucleus |
| ubiquitin ligase complex |
| Function |
| catalytic activity |
| small conjugating protein ligase activity |
| ligase activity |
| ligase activity, forming carbon-nitrogen bonds |
| acid-amino acid ligase activity |
| Molecular Function |
| acid-amino acid ligase activity |
| ubiquitin-protein ligase activity |
| ATP binding |
| transcription coactivator activity |
| enzyme binding |
| Process |
| metabolic process |
| regulation of protein metabolic process |
| macromolecule metabolic process |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| post-translational protein modification |
| macromolecule modification |
| protein modification process |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protein ubiquitination | Not Available | Not Available | | Ubiquitin mediated proteolysis | Not Available | 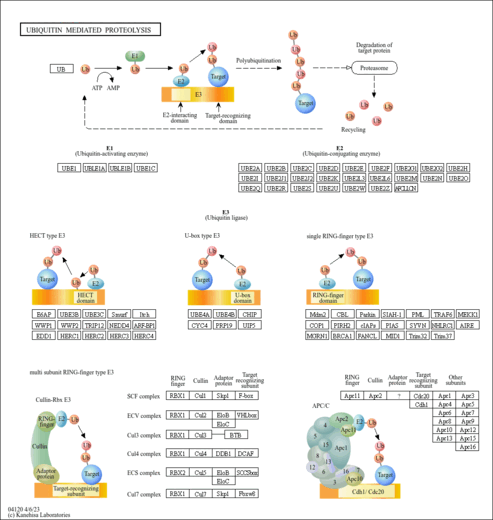 | | Parkinson's disease | Not Available | 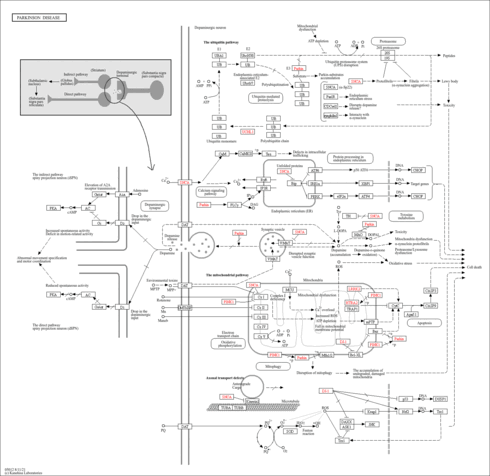 |
|
| Gene Properties |
|---|
| Chromosome Location
| 22 |
| Locus
| 22q11.21 |
| SNPs
| UBE2L3 |
| Gene Sequence
|
>465 bp
ATGGCGGCCAGCAGGAGGCTGATGAAGGAGCTTGAAGAAATCCGCAAATGTGGGATGAAA
AACTTCCGTAACATCCAGGTTGATGAAGCTAATTTATTGACTTGGCAAGGGCTTATTGTT
CCTGACAACCCTCCATATGATAAGGGAGCCTTCAGAATCGAAATCAACTTTCCAGCAGAG
TACCCATTCAAACCACCGAAGATCACATTTAAAACAAAGATCTATCACCCAAACATCGAC
GAAAAGGGGCAGGTCTGTCTGCCAGTAATTAGTGCCGAAAACTGGAAGCCAGCAACCAAA
ACCGACCAAGTAATCCAGTCCCTCATAGCACTGGTGAATGACCCCCAGCCTGAGCACCCG
CTTCGGGCTGACCTAGCTGAAGAATACTCTAAGGACCGTAAAAAATTCTGTAAGAATGCT
GAAGAGTTTACAAAGAAATATGGGGAAAAGCGACCTGTGGACTAA
|
| Protein Properties |
|---|
| Number of Residues
| 154 |
| Molecular Weight
| 24003.385 |
| Theoretical pI
| 8.742 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ubiquitin-conjugating enzyme E2 L3
MAASRRLMKELEEIRKCGMKNFRNIQVDEANLLTWQGLIVPDNPPYDKGAFRIEINFPAE
YPFKPPKITFKTKIYHPNIDEKGQVCLPVISAENWKPATKTDQVIQSLIALVNDPQPEHP
LRADLAEEYSKDRKKFCKNAEEFTKKYGEKRPVD
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| P68036 |
| UniProtKB/Swiss-Prot Entry Name
| UB2L3_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| S81003 |
| GeneCard ID
| UBE2L3 |
| GenAtlas ID
| UBE2L3 |
| HGNC ID
| HGNC:12488 |
| References |
|---|
| General References
| Not Available |

