| Identification |
|---|
| HMDB Protein ID
| CDBP00423 |
| Secondary Accession Numbers
| Not Available |
| Name
| Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- dUTP pyrophosphatase
- dUTPase
|
| Gene Name
| DUT |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity |
| Specific Function
| This enzyme is involved in nucleotide metabolism: it produces dUMP, the immediate precursor of thymidine nucleotides and it decreases the intracellular concentration of dUTP so that uracil cannot be incorporated into DNA.
|
| GO Classification
|
| Biological Process |
| pyrimidine nucleoside biosynthetic process |
| pyrimidine nucleobase metabolic process |
| DNA replication |
| dUMP biosynthetic process |
| dUTP metabolic process |
| Cellular Component |
| mitochondrion |
| nucleoplasm |
| Function |
| catalytic activity |
| hydrolase activity |
| nucleoside-triphosphate diphosphatase activity |
| dutp diphosphatase activity |
| hydrolase activity, acting on acid anhydrides |
| hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides |
| pyrophosphatase activity |
| Molecular Function |
| dUTP diphosphatase activity |
| Process |
| nucleotide metabolic process |
| pyrimidine nucleoside triphosphate metabolic process |
| pyrimidine deoxyribonucleoside triphosphate metabolic process |
| dutp metabolic process |
| pyrimidine nucleotide metabolic process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
|
| Cellular Location
|
- Isoform 2:Nucleus
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | dUMP biosynthesis | Not Available | Not Available | | Pyrimidine Metabolism |  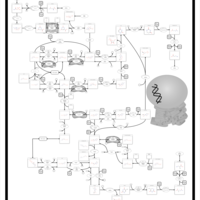  |  | | Beta Ureidopropionase Deficiency |  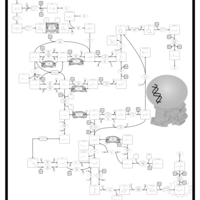 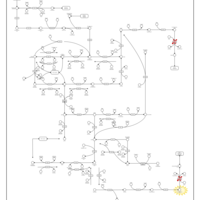 | Not Available | | UMP Synthase Deficiency (Orotic Aciduria) |  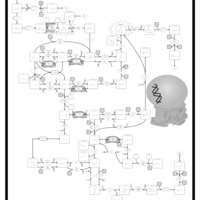  | Not Available | | Dihydropyrimidinase Deficiency |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 15 |
| Locus
| 15q21.1 |
| SNPs
| DUT |
| Gene Sequence
|
>759 bp
ATGACTCCCCTCTGCCCTCGCCCCGCGCTCTGCTACCATTTCCTTACGTCTCTGCTTCGC
TCAGCGATGCAAAACGCGCGAGGCGCACGGCAGAGGGCCGAAGCCGCGGTACTCTCCGGG
CCAGGCCCGCCCCTCGGCCGCGCCGCGCAGCACGGGATTCCCCGGCCGCTGTCCAGCGCT
GGCCGCCTGAGCCAAGGCTGCCGCGGAGCCAGTACAGTCGGGGCCGCTGGCTGGAAGGGC
GAGCTTCCTAAGGCGGGGGGAAGCCCGGCGCCGGGGCCGGAGACACCCGCCATTTCACCC
AGTAAGCGGGCCCGGCCTGCGGAGGTGGGCGGCATGCAGCTCCGCTTTGCCCGGCTCTCC
GAGCACGCCACGGCCCCCACCCGGGGCTCCGCGCGCGCCGCGGGCTACGACCTGTACAGT
GCCTATGATTACACAATACCACCTATGGAGAAAGCTGTTGTGAAAACGGACATTCAGATA
GCGCTCCCTTCTGGGTGTTATGGAAGAGTGGCTCCACGGTCAGGCTTGGCTGCAAAACAC
TTTATTGATGTAGGAGCTGGTGTCATAGATGAAGATTATAGAGGAAATGTTGGTGTTGTA
CTGTTTAATTTTGGCAAAGAAAAGTTTGAAGTCAAAAAAGGTGATCGAATTGCACAGCTC
ATTTGCGAACGGATTTTTTATCCAGAAATAGAAGAAGTTCAAGCCTTGGATGACACCGAA
AGGGGTTCAGGAGGTTTTGGTTCCACTGGAAAGAATTAA
|
| Protein Properties |
|---|
| Number of Residues
| 252 |
| Molecular Weight
| 26562.975 |
| Theoretical pI
| 9.368 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial
MTPLCPRPALCYHFLTSLLRSAMQNARGTAEGRSRGTLRARPAPRPPAAQHGIPRPLSSA
GRLSQGCRGASTVGAAGWKGELPKAGGSPAPGPETPAISPSKRARPAEVGGMQLRFARLS
EHATAPTRGSARAAGYDLYSAYDYTIPPMEKAVVKTDIQIALPSGCYGRVAPRSGLAAKH
FIDVGAGVIDEDYRGNVGVVLFNFGKEKFEVKKGDRIAQLICERIFYPEIEEVQALDDTE
RGSGGFGSTGKN
|
| External Links |
|---|
| GenBank ID Protein
| 70906441 |
| UniProtKB/Swiss-Prot ID
| P33316 |
| UniProtKB/Swiss-Prot Entry Name
| DUT_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_001025248.1 |
| GeneCard ID
| DUT |
| GenAtlas ID
| DUT |
| HGNC ID
| HGNC:3078 |
| References |
|---|
| General References
| Not Available |
