| Identification |
|---|
| HMDB Protein ID
| CDBP00396 |
| Secondary Accession Numbers
| Not Available |
| Name
| Histone-lysine N-methyltransferase SETD7 |
| Description
| Not Available |
| Synonyms
|
- H3-K4-HMTase SETD7
- Histone H3-K4 methyltransferase SETD7
- Lysine N-methyltransferase 7
- SET domain-containing protein 7
- SET7/9
|
| Gene Name
| SETD7 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in histone-lysine N-methyltransferase activity |
| Specific Function
| Histone methyltransferase that specifically monomethylates 'Lys-4' of histone H3. H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. Plays a central role in the transcriptional activation of genes such as collagenase or insulin. Recruited by IPF1/PDX-1 to the insulin promoter, leading to activate transcription. Has also methyltransferase activity toward non-histone proteins such as p53/TP53, TAF10, and possibly TAF7 by recognizing and binding the [KR]-[STA]-K in substrate proteins. Monomethylates 'Lys-189' of TAF10, leading to increase the affinity of TAF10 for RNA polymerase II. Monomethylates 'Lys-372' of p53/TP53, stabilizing p53/TP53 and increasing p53/TP53-mediated transcriptional activation.
|
| GO Classification
|
| Biological Process |
| peptidyl-lysine dimethylation |
| peptidyl-lysine monomethylation |
| regulation of transcription, DNA-dependent |
| transcription, DNA-dependent |
| Cellular Component |
| chromosome |
| nucleus |
| Component |
| organelle |
| non-membrane-bounded organelle |
| intracellular non-membrane-bounded organelle |
| chromosome |
| Function |
| methyltransferase activity |
| catalytic activity |
| transferase activity |
| protein methyltransferase activity |
| protein-lysine n-methyltransferase activity |
| histone-lysine n-methyltransferase activity |
| transferase activity, transferring one-carbon groups |
| Molecular Function |
| histone-lysine N-methyltransferase activity |
| Process |
| cellular process |
| cellular component organization at cellular level |
| organelle organization |
| chromosome organization |
| chromatin organization |
| chromatin modification |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| regulation of gene expression |
| regulation of transcription |
| regulation of transcription, dna-dependent |
|
| Cellular Location
|
- Nucleus
- Chromosome (Probable)
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Carnitine Synthesis | 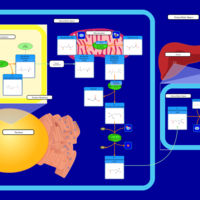 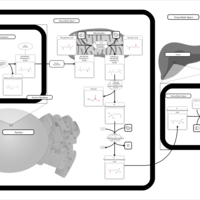 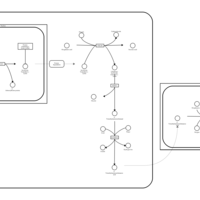 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 4 |
| Locus
| 4q28 |
| SNPs
| SETD7 |
| Gene Sequence
|
>1101 bp
ATGGATAGCGACGACGAGATGGTGGAGGAGGCGGTGGAAGGGCACCTGGACGATGACGGA
TTACCGCACGGGTTCTGCACAGTCACCTACTCCTCCACAGACAGATTTGAGGGGAACTTT
GTTCACGGAGAAAAGAACGGACGGGGGAAGTTCTTCTTCTTTGATGGCAGCACCCTGGAG
GGGTATTATGTGGATGATGCCTTGCAGGGCCAGGGAGTTTACACTTACGAAGATGGGGGA
GTTCTCCAGGGCACGTATGTAGACGGAGAGCTGAACGGTCCAGCCCAGGAATATGACACA
GATGGGAGACTGATCTTCAAGGGGCAGTATAAAGATAACATTCGTCATGGAGTGTGCTGG
ATATATTACCCAGATGGAGGAAGCCTTGTAGGAGAAGTAAATGAAGATGGGGAGATGACT
GGAGAGAAGATAGCCTATGTGTACCCTGATGAGAGGACCGCACTTTATGGGAAATTTATT
GATGGAGAGATGATAGAAGGCAAACTGGCTACCCTTATGTCCACTGAAGAAGGGAGGCCT
CACTTTGAACTGATGCCTGGAAATTCAGTGTACCACTTTGATAAGTCGACTTCATCTTGC
ATTTCTACCAATGCTCTTCTTCCAGATCCTTATGAATCAGAAAGGGTTTATGTTGCTGAA
TCTCTTATTTCCAGTGCTGGAGAAGGACTTTTTTCAAAGGTAGCTGTGGGACCTAATACT
GTTATGTCTTTTTATAATGGAGTTCGAATTACACACCAAGAGGTTGACAGCAGGGACTGG
GCCCTTAATGGGAACACCCTCTCCCTTGATGAAGAAACGGTCATTGATGTGCCTGAGCCC
TATAACCACGTATCCAAGTACTGTGCCTCCTTGGGACACAAGGCAAATCACTCCTTCACT
CCAAACTGCATCTACGATATGTTTGTCCACCCCCGTTTTGGGCCCATCAAATGCATCCGC
ACCCTGAGAGCAGTGGAGGCCGATGAAGAGCTCACCGTTGCCTATGGCTATGACCACAGC
CCCCCCGGGAAGAGTGGGCCTGAAGCCCCTGAGTGGTACCAGGTGGAGCTGAAGGCCTTC
CAGGCCACCCAGCAAAAGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 366 |
| Molecular Weight
| 40720.595 |
| Theoretical pI
| 4.625 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Histone-lysine N-methyltransferase SETD7
MDSDDEMVEEAVEGHLDDDGLPHGFCTVTYSSTDRFEGNFVHGEKNGRGKFFFFDGSTLE
GYYVDDALQGQGVYTYEDGGVLQGTYVDGELNGPAQEYDTDGRLIFKGQYKDNIRHGVCW
IYYPDGGSLVGEVNEDGEMTGEKIAYVYPDERTALYGKFIDGEMIEGKLATLMSTEEGRP
HFELMPGNSVYHFDKSTSSCISTNALLPDPYESERVYVAESLISSAGEGLFSKVAVGPNT
VMSFYNGVRITHQEVDSRDWALNGNTLSLDEETVIDVPEPYNHVSKYCASLGHKANHSFT
PNCIYDMFVHPRFGPIKCIRTLRAVEADEELTVAYGYDHSPPGKSGPEAPEWYQVELKAF
QATQQK
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q8WTS6 |
| UniProtKB/Swiss-Prot Entry Name
| SETD7_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF448510 |
| GeneCard ID
| SETD7 |
| GenAtlas ID
| SETD7 |
| HGNC ID
| HGNC:30412 |
| References |
|---|
| General References
| Not Available |