| Identification |
|---|
| HMDB Protein ID
| CDBP00384 |
| Secondary Accession Numbers
| Not Available |
| Name
| Lactoylglutathione lyase |
| Description
| Not Available |
| Synonyms
|
- Aldoketomutase
- Glx I
- Glyoxalase I
- Ketone-aldehyde mutase
- Methylglyoxalase
- S-D-lactoylglutathione methylglyoxal lyase
|
| Gene Name
| GLO1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in lactoylglutathione lyase activity |
| Specific Function
| Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione. Involved in the regulation of TNF-induced transcriptional activity of NF-kappa-B.
|
| GO Classification
|
| Biological Process |
| negative regulation of apoptotic process |
| methylglyoxal metabolic process |
| glutathione metabolic process |
| regulation of transcription from RNA polymerase II promoter |
| carbohydrate metabolic process |
| Cellular Component |
| cytoplasm |
| Function |
| carbon-sulfur lyase activity |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| lyase activity |
| lactoylglutathione lyase activity |
| Molecular Function |
| metal ion binding |
| lactoylglutathione lyase activity |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Pyruvaldehyde Degradation | 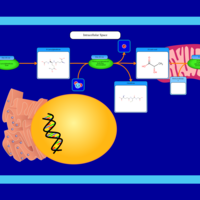 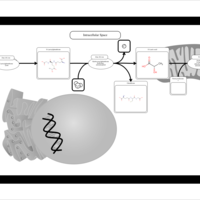 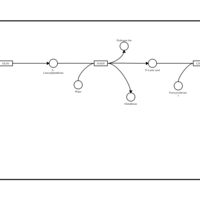 | Not Available | | Pyruvate Metabolism |  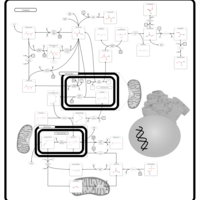  |  | | methylglyoxal degradation | Not Available | Not Available | | Leigh Syndrome | 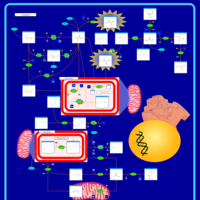 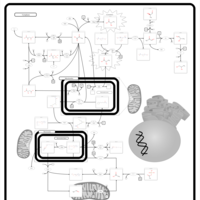 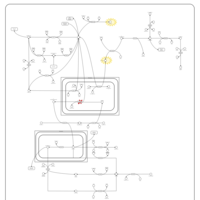 | Not Available | | Pyruvate Decarboxylase E1 Component Deficiency (PDHE1 Deficiency) | 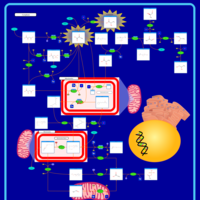   | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 6 |
| Locus
| 6p21.3-p21.1 |
| SNPs
| GLO1 |
| Gene Sequence
|
>555 bp
ATGGCAGAACCGCAGCCCCCGTCCGGCGGCCTCACGGACGAGGCCGCCCTCAGTTGCTGC
TCCGACGCGGACCCCAGTACCAAGGATTTTCTATTGCAGCAGACCATGCTACGAGTGAAG
GATCCTAAGAAGTCACTGGATTTTTATACTAGAGTTCTTGGAATGACGCTAATCCAAAAA
TGTGATTTTCCCATTATGAAGTTTTCACTCTACTTCTTGGCTTATGAGGATAAAAATGAC
ATCCCTAAAGAAAAAGATGAAAAAATAGCCTGGGCGCTCTCCAGAAAAGCTACACTTGAG
CTGACACACAATTGGGGCACTGAAGATGATGCGACCCAGAGTTACCACAATGGCAATTCA
GACCCTCGAGGATTCGGTCATATTGGAATTGCTGTTCCTGATGTATACAGTGCTTGTAAA
AGGTTTGAAGAACTGGGAGTCAAATTTGTGAAGAAACCTGATGATGGTAAAATGAAAGGC
CTGGCATTTATTCAAGATCCTGATGGCTACTGGATTGAAATTTTGAATCCTAACAAAATG
GCAACCTTAATGTAG
|
| Protein Properties |
|---|
| Number of Residues
| 184 |
| Molecular Weight
| 20777.515 |
| Theoretical pI
| 5.309 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Lactoylglutathione lyase
MAEPQPPSGGLTDEAALSCCSDADPSTKDFLLQQTMLRVKDPKKSLDFYTRVLGMTLIQK
CDFPIMKFSLYFLAYEDKNDIPKEKDEKIAWALSRKATLELTHNWGTEDDETQSYHNGNS
DPRGFGHIGIAVPDVYSACKRFEELGVKFVKKPDDGKMKGLAFIQDPDGYWIEILNPNKM
ATLM
|
| External Links |
|---|
| GenBank ID Protein
| 5020074 |
| UniProtKB/Swiss-Prot ID
| Q04760 |
| UniProtKB/Swiss-Prot Entry Name
| LGUL_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF146651 |
| GeneCard ID
| GLO1 |
| GenAtlas ID
| GLO1 |
| HGNC ID
| HGNC:4323 |
| References |
|---|
| General References
| Not Available |
