| Identification |
|---|
| HMDB Protein ID
| CDBP00373 |
| Secondary Accession Numbers
| Not Available |
| Name
| Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- HCDH
- Medium and short-chain L-3-hydroxyacyl-coenzyme A dehydrogenase
- Short-chain 3-hydroxyacyl-CoA dehydrogenase
|
| Gene Name
| HADH |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in 3-hydroxyacyl-CoA dehydrogenase activity |
| Specific Function
| Plays an essential role in the mitochondrial beta-oxidation of short chain fatty acids. Exerts it highest activity toward 3-hydroxybutyryl-CoA.
|
| GO Classification
|
| Biological Process |
| response to insulin stimulus |
| response to drug |
| negative regulation of insulin secretion |
| response to activity |
| fatty acid beta-oxidation |
| Cellular Component |
| mitochondrial matrix |
| nucleus |
| mitochondrial inner membrane |
| Function |
| coenzyme binding |
| cofactor binding |
| nad binding |
| 3-hydroxyacyl-coa dehydrogenase activity |
| oxidoreductase activity, acting on ch-oh group of donors |
| oxidoreductase activity |
| binding |
| nucleotide binding |
| catalytic activity |
| nad or nadh binding |
| oxidoreductase activity, acting on the ch-oh group of donors, nad or nadp as acceptor |
| Molecular Function |
| 3-hydroxyacyl-CoA dehydrogenase activity |
| NAD+ binding |
| Process |
| metabolic process |
| cellular metabolic process |
| organic acid metabolic process |
| oxoacid metabolic process |
| carboxylic acid metabolic process |
| monocarboxylic acid metabolic process |
| fatty acid metabolic process |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | fatty acid beta-oxidation | Not Available | Not Available | | Fatty acid elongation | Not Available | 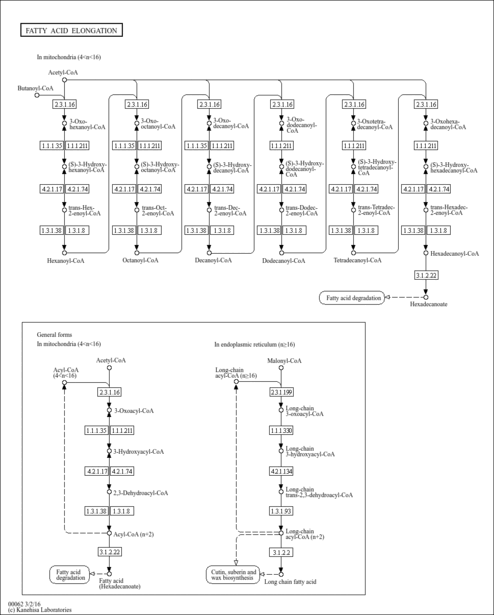 | | Butanoate metabolism | Not Available |  | | Butyrate Metabolism |    |  | | Lysine Degradation | 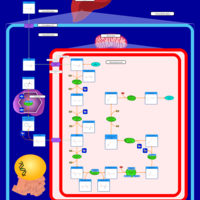 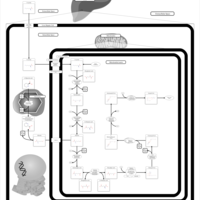 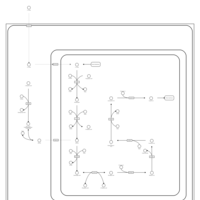 |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 4 |
| Locus
| 4q22-q26 |
| SNPs
| HADH |
| Gene Sequence
|
>945 bp
ATGGCCTTCGTCACCAGGCAGTTCATGCGTTCCGTGTCCTCCTCGTCCACCGCCTCGGCC
TCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCC
GGCATTGCCCAGGTTGCTGCAGCAACTGGTCACACAGTAGTGTTGGTAGACCAGACAGAG
GACATCCTGGCAAAATCCAAAAAGGGAATTGAGGAAAGCCTTAGGAAAGTGGCAAAGAAG
AAGTTTGCAGAAAACCCTAAGGCCGGCGATGAATTTGTGGAGAAGACCCTGAGCACCATA
GCGACCAGCACGGATGCAGCCTCCGTTGTCCACAGCACAGACTTGGTGGTGGAAGCCATC
GTGGAGAATCTGAAGGTGAAAAACGAGCTCTTCAAAAGGCTGGACAAGTTTGCTGCTGAA
CATACAATCTTTGCCAGCAACACTTCCTCCTTGCATATTACAAGCATAGCTAATGCCACC
ACCAGACAAGACCGATTCGCTGGCCTCCATTTCTTCAACCCAGTGCCTGTCATGAAACTT
GTGGAGGTCATTAAAACACCAATGACCAGCCAGAAGACATTTGAATCTTTGGTAGACTTT
AGCAAAGCCCTAGGAAAGCATCCTGTTTCTTGCAAGGACACTCCTGGGTTTATTGTGAAC
CGCCTCCTGGTTCCATACCTCATGGAAGCAATCAGGCTGTATGAACGAGGTGACGCATCC
AAAGAAGACATTGACACTGCTATGAAATTAGGAGCCGGTTACCCCATGGGCCCATTTGAG
CTTCTAGATTATGTCGGACTGGATACTACGAAGTTCATCGTGGATGGGTGGCATGAAATG
GATGCAGAGAACCCATTACATCAGCCCAGCCCATCCTTAAATAAGCTGGTAGCAGAGAAC
AAGTTCGGCAAGAAGACTGGAGAAGGATTTTACAAATACAAGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 314 |
| Molecular Weight
| 36035.11 |
| Theoretical pI
| 8.535 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial
MAFVTRQFMRSVSSSSTASASAKKIIVKHVTVIGGGLMGAGIAQVAAATGHTVVLVDQTE
DILAKSKKGIEESLRKVAKKKFAENPKAGDEFVEKTLSTIATSTDAASVVHSTDLVVEAI
VENLKVKNELFKRLDKFAAEHTIFASNTSSLQITSIANATTRQDRFAGLHFFNPVPVMKL
VEVIKTPMTSQKTFESLVDFSKALGKHPVSCKDTPGFIVNRLLVPYLMEAIRLYERGDAS
KEDIDTAMKLGAGYPMGPFELLDYVGLDTTKFIVDGWHEMDAENPLHQPSPSLNKLVAEN
KFGKKTGEGFYKYK
|
| External Links |
|---|
| GenBank ID Protein
| 1483511 |
| UniProtKB/Swiss-Prot ID
| Q16836 |
| UniProtKB/Swiss-Prot Entry Name
| HCDH_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| X96752 |
| GeneCard ID
| HADH |
| GenAtlas ID
| HADH |
| HGNC ID
| HGNC:4799 |
| References |
|---|
| General References
| Not Available |


