| Identification |
|---|
| HMDB Protein ID
| CDBP00201 |
| Secondary Accession Numbers
| Not Available |
| Name
| D-amino-acid oxidase |
| Description
| Not Available |
| Synonyms
|
- DAAO
- DAMOX
- DAO
|
| Gene Name
| DAO |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in D-amino-acid oxidase activity |
| Specific Function
| Regulates the level of the neuromodulator D-serine in the brain. Has high activity towards D-DOPA and contributes to dopamine synthesis. Could act as a detoxifying agent which removes D-amino acids accumulated during aging. Acts on a variety of D-amino acids with a preference for those having small hydrophobic side chains followed by those bearing polar, aromatic, and basic groups. Does not act on acidic amino acids.
|
| GO Classification
|
| Biological Process |
| cellular nitrogen compound metabolic process |
| glyoxylate metabolic process |
| dopamine biosynthetic process |
| D-alanine catabolic process |
| D-serine catabolic process |
| leucine metabolic process |
| proline catabolic process |
| Cellular Component |
| cytosol |
| peroxisomal matrix |
| peroxisomal membrane |
| Function |
| catalytic activity |
| oxidoreductase activity, acting on the ch-nh2 group of donors |
| oxidoreductase activity, acting on the ch-nh2 group of donors, oxygen as acceptor |
| d-amino-acid oxidase activity |
| oxidoreductase activity |
| binding |
| Molecular Function |
| D-amino-acid oxidase activity |
| FAD binding |
| protein dimerization activity |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Peroxisome
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glycine, serine and threonine metabolism | Not Available |  | | Peroxisome | Not Available | 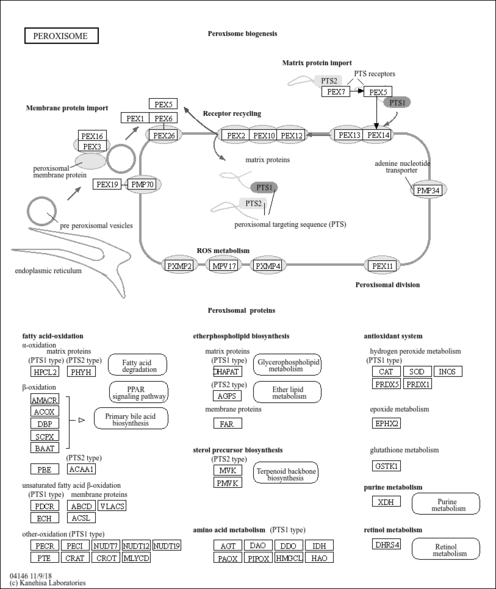 | | D-Arginine and D-Ornithine Metabolism |    | 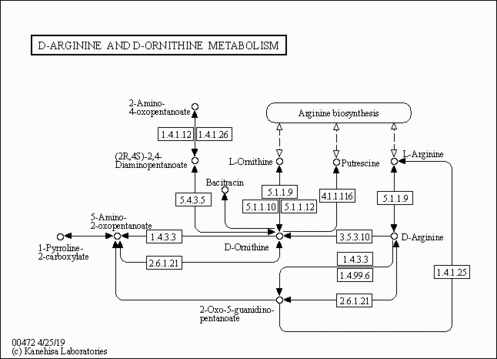 | | Arginine and Proline Metabolism |    |  | | Prolidase Deficiency (PD) | 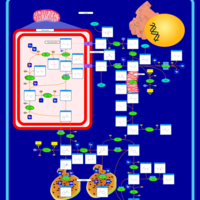 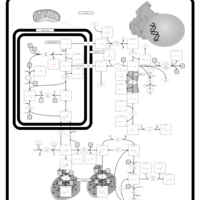 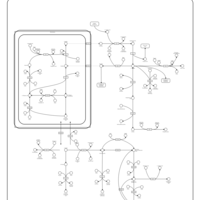 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 12 |
| Locus
| 12q24 |
| SNPs
| DAO |
| Gene Sequence
|
>1044 bp
ATGCGTGTGGTGGTGATTGGAGCAGGAGTCATCGGGCTGTCCACCGCCCTCTGCATCCAT
GAGCGCTACCACTCAGTCCTGCAGCCACTGCACATAAAGGTCTACGCGGACCGCTTCACC
CCACTCACCACCACCGACGTGGCTGCCGGCCTCTGGCAGCCCTACCTTTCTGACCCCAAC
AACCCACAGGAGGCGGACTGGAGCCAACAGACCTTTGACTATCTCCTGAGCCATGTCCAT
TCTCCCAACGCTGAAAACCTGGGCCTGTTCCTAATCTCGGGCTACAACCTCTTCCATGAA
GCCATTCCGGACCCTTCCTGGAAGGACACAGTTCTGGGATTTCGGAAGCTGACCCCCAGA
GAGCTGGATATGTTCCCAGATTACGGCTATGGCTGGTTCCACACAAGCCTAATTCTGGAG
GGAAAGAACTATCTACAGTGGCTGACTGAAAGGTTAACTGAGAGGGGAGTGAAGTTCTTC
CAGCGGAAAGTGGAGTCTTTTGAGGAGGTGGCAAGAGAAGGCGCAGACGTGATTGTCAAC
TGCACTGGGGTATGGGCTGGGGCGCTACAACGAGACCCCCTGCTGCAGCCAGGCCGGGGG
CAGATCATGAAGGTGGACGCCCCTTGGATGAAGCACTTCATTCTCACCCATGACCCAGAG
AGAGGCATCTACAATTCCCCGTACATCATCCCAGGGACCCAGACAGTTACTCTTGGAGGC
ATCTTCCAGTTGGGAAACTGGAGTGAACTAAACAATATCCAGGACCACAACACCATTTGG
GAAGGCTGCTGCAGACTGGAGCCCACACTGAAGAATGCAAGAATTATTGGTGAAGCAACT
GGCTTCCGGCCAGTACGCCCCCAGATTCGGCTAGAAAGAGAACAGCTTCGCACTGGACCT
TCAAACACAGAGGTCATCCACAACTATGGCCATGGAGGCTACGGGCTCACCATCCACTGG
GGATGTGCCCTGGAGGCAGCCAAGCTCTTTGGGAGAATCCTGGAAGAAAAGAAATTGTCC
AGAATGCCACCATCCCACCTCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 347 |
| Molecular Weight
| 39473.75 |
| Theoretical pI
| 6.844 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>D-amino-acid oxidase
MRVVVIGAGVIGLSTALCIHERYHSVLQPLDIKVYADRFTPLTTTDVAAGLWQPYLSDPN
NPQEADWSQQTFDYLLSHVHSPNAENLGLFLISGYNLFHEAIPDPSWKDTVLGFRKLTPR
ELDMFPDYGYGWFHTSLILEGKNYLQWLTERLTERGVKFFQRKVESFEEVAREGADVIVN
CTGVWAGALQRDPLLQPGRGQIMKVDAPWMKHFILTHDPERGIYNSPYIIPGTQTVTLGG
IFQLGNWSELNNIQDHNTIWEGCCRLEPTLKNARIIGERTGFRPVRPQIRLEREQLRTGP
SNTEVIHNYGHGGYGLTIHWGCALEAAKLFGRILEEKKLSRMPPSHL
|
| External Links |
|---|
| GenBank ID Protein
| 30446 |
| UniProtKB/Swiss-Prot ID
| P14920 |
| UniProtKB/Swiss-Prot Entry Name
| OXDA_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| X13227 |
| GeneCard ID
| DAO |
| GenAtlas ID
| DAO |
| HGNC ID
| HGNC:2671 |
| References |
|---|
| General References
| Not Available |



