| Identification |
|---|
| HMDB Protein ID
| CDBP00200 |
| Secondary Accession Numbers
| Not Available |
| Name
| Retinal dehydrogenase 2 |
| Description
| Not Available |
| Synonyms
|
- Aldehyde dehydrogenase family 1 member A2
- RALDH 2
- RALDH(II)
- RalDH2
- Retinaldehyde-specific dehydrogenase type 2
|
| Gene Name
| ALDH1A2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Recognizes as substrates free retinal and cellular retinol-binding protein-bound retinal. Does metabolize octanal and decanal but does not metabolize citral, benzaldehyde, acetaldehyde and propanal efficiently (By similarity).
|
| GO Classification
|
| Biological Process |
| negative regulation of cell proliferation |
| retinal metabolic process |
| neural crest cell development |
| 9-cis-retinoic acid biosynthetic process |
| neural tube development |
| anterior/posterior pattern specification |
| neuron differentiation |
| blood vessel development |
| pancreas development |
| cellular response to retinoic acid |
| pituitary gland development |
| determination of bilateral symmetry |
| positive regulation of gene expression |
| embryonic camera-type eye development |
| proximal/distal pattern formation |
| embryonic forelimb morphogenesis |
| regulation of endothelial cell proliferation |
| face development |
| response to cytokine stimulus |
| embryonic digestive tract development |
| response to estradiol stimulus |
| heart morphogenesis |
| positive regulation of apoptotic process |
| response to vitamin A |
| hindbrain development |
| positive regulation of cell proliferation |
| retinoic acid receptor signaling pathway |
| kidney development |
| retinol metabolic process |
| ureter maturation |
| lung development |
| retinoic acid metabolic process |
| vitamin A metabolic process |
| midgut development |
| morphogenesis of embryonic epithelium |
| liver development |
| Cellular Component |
| cytoplasm |
| nucleus |
| cytosol |
| Function |
| catalytic activity |
| oxidoreductase activity |
| Molecular Function |
| retinal dehydrogenase activity |
| 3-chloroallyl aldehyde dehydrogenase activity |
| retinal binding |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Retinol Metabolism | 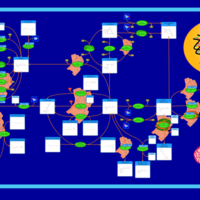 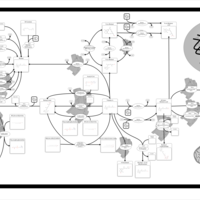 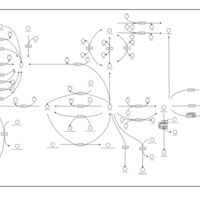 | 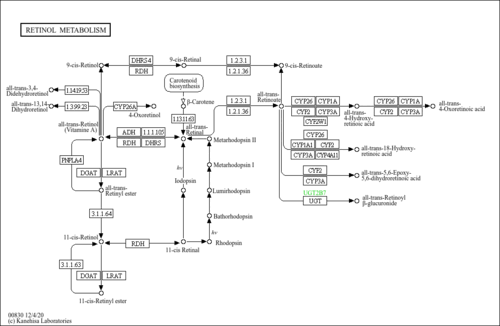 | | Vitamin A Deficiency | 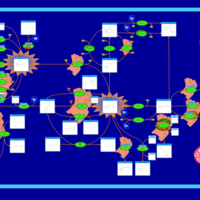 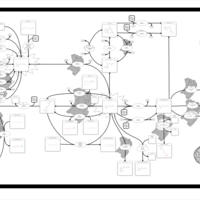 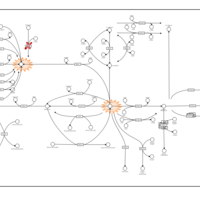 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 15 |
| Locus
| 15q21.3 |
| SNPs
| ALDH1A2 |
| Gene Sequence
|
>1557 bp
ATGACTTCCAGCAAGATAGAGATGCCCGGCGAGGTGAAGGCCGACCCCGCCGCCCTCATG
GCGTCGCTGCACCTCCTGCCGTCGCCCACGCCCAATCTCGAAATTAAGTACACCAAGATC
TTTATAAACAACGAGTGGCAGAACTCAGAGAGTGGGAGAGTGTTCCCTGTCTATAATCCA
GCCACAGGAGAACAGGTGTGTGAAGTTCAAGAAGCAGACAAGGCAGATATAGACAAAGCA
GTGCAGGCAGCCCGCCTGGCTTTCTCTCTTGGTTCAGTGTGGAGAAGGATGGATGCTTCA
GAAAGGGGACGTCTGTTGGATAAGCTTGCAGACTTGGTGGAACGGGACAGGGCAGTTCTT
GCAACCATGGAATCCCTAAATGGTGGCAAACCATTCCTGCAAGCTTTTTATGTGGATTTG
CAGGGCGTCATCAAAACCTTTCGATATTACGCAGGCTGGGCTGATAAAATTCATGGGATG
ACCATTCCTGTAGATGGAGACTATTTTACCTTTACAAGACATGAACCCATTGGAGTGTGT
GGACAGATCATCCCATGGAACTTCCCCCTGCTGATGTTTGCCTGGAAAATAGCTCCAGCT
TTGTGCTGTGGCAATACAGTAGTTATTAAGCCAGCAGAGCAAACACCACTCAGTGCACTC
TACATGGGAGCCCTCATCAAGGAGGCTGGCTTTCCTCCCGGGGTCATCAATATTTTGCCA
GGATATGGGCCAACGGCTGGGGCAGCAATAGCTTCTCACATTGGCATAGACAAGATTGCA
TTCACAGGGTCTACTGAGGTTGGAAAGCTTATCCAAGAAGCAGCTGGAAGAAGTAATTTG
AAGAGAGTAACTCTGGAACTTGGAGGCAAAAGTCCTAATATTATTTTTGCTGATGCTGAC
TTGGACTATGCTGTGGAGCAGGCCCACCAGGGTGTGTTCTTCAATCAAGGTCAGTGCTGC
ACTGCAGGCTCTCGCATCTTCGTGGAGGAGTCCATCTATGAGGAGTTTGTGAGAAGAAGC
GTGGAGCGGGCCAAGAGGCGCATAGTGGGGAGTCCCTTTGACCCCACCACTGAGCAGGGT
CCCCAGATTGATAAGAAACAGTACAACAAGATCTTGGAACTCATCCAGAGTGGTGTGGCT
GAGGGCGCCAAGCTGGAATGTGGAGGCAAAGGACTGGGCCGAAAGGGGTTTTTCATTGAG
CCCACAGTGTTTTCCAACGTCACTGATGATATGCGGATTGCCAAGGAGGAGATCTTTGGC
CCTGTTCAGGAAATTTTGAGATTTAAGACGATGGATGAAGTTATCGAAAGAGCCAATAAC
TCAGACTTTGGACTCGTAGCAGCTGTCTTTACTAATGACATCAACAAGGCCCTCACAGTG
TCTTCTGCAATGCAAGCTGGGACTGTTTGGATCAATTGTTACAATGCCTTAAATGCCCAG
AGCCCCTTTGGGGGATTCAAGATGTCTGGAAATGGGAGAGAAATGGGAGAATTTGGCTTG
CGGGAGTACTCAGAAGTTAAGACGGTGACAGTAAAGATCCCCCAGAAGAACTCCTAA
|
| Protein Properties |
|---|
| Number of Residues
| 518 |
| Molecular Weight
| 54672.24 |
| Theoretical pI
| 6.589 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Retinal dehydrogenase 2
MTSSKIEMPGEVKADPAALMASLHLLPSPTPNLEIKYTKIFINNEWQNSESGRVFPVYNP
ATGEQVCEVQEADKADIDKAVQAARLAFSLGSVWRRMDASERGRLLDKLADLVERDRAVL
ATMESLNGGKPFLQAFYVDLQGVIKTFRYYAGWADKIHGMTIPVDGDYFTFTRHEPIGVC
GQIIPWNFPLLMFAWKIAPALCCGNTVVIKPAEQTPLSALYMGALIKEAGFPPGVINILP
GYGPTAGAAIASHIGIDKIAFTGSTEVGKLIQEAAGRSNLKRVTLELGGKSPNIIFADAD
LDYAVEQAHQGVFFNQGQCCTAGSRIFVEESIYEEFVRRSVERAKRRVVGSPFDPTTEQG
PQIDKKQYNKILELIQSGVAEGAKLECGGKGLGRKGFFIEPTVFSNVTDDMRIAKEEIFG
PVQEILRFKTMDEVIERANNSDFGLVAAVFTNDINKALTVSSAMQAGTVWINCYNALNAQ
SPFGGFKMSGNGREMGEFGLREYSEVKTVTVKIPQKNS
|
| External Links |
|---|
| GenBank ID Protein
| 3970842 |
| UniProtKB/Swiss-Prot ID
| O94788 |
| UniProtKB/Swiss-Prot Entry Name
| AL1A2_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AB015226 |
| GeneCard ID
| ALDH1A2 |
| GenAtlas ID
| ALDH1A2 |
| HGNC ID
| HGNC:15472 |
| References |
|---|
| General References
| Not Available |
