| Identification |
|---|
| HMDB Protein ID
| CDBP00053 |
| Secondary Accession Numbers
| Not Available |
| Name
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- 70 kDa mitochondrial autoantigen of primary biliary cirrhosis
- Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex
- M2 antigen complex 70 kDa subunit
- PBC
- PDC-E2
- PDCE2
- Pyruvate dehydrogenase complex component E2
|
| Gene Name
| DLAT |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acyltransferase activity |
| Specific Function
| The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2), and thereby links the glycolytic pathway to the tricarboxylic cycle.
|
| GO Classification
|
| Biological Process |
| regulation of acetyl-CoA biosynthetic process from pyruvate |
| sleep |
| glycolysis |
| pyruvate metabolic process |
| acetyl-CoA biosynthetic process |
| acetyl-CoA biosynthetic process from pyruvate |
| Cellular Component |
| mitochondrial pyruvate dehydrogenase complex |
| Component |
| macromolecular complex |
| protein complex |
| pyruvate dehydrogenase complex |
| Function |
| transferase activity, transferring acyl groups other than amino-acyl groups |
| acyltransferase activity |
| acetyltransferase activity |
| protein binding |
| s-acetyltransferase activity |
| dihydrolipoyllysine-residue acetyltransferase activity |
| binding |
| catalytic activity |
| transferase activity |
| transferase activity, transferring acyl groups |
| Molecular Function |
| dihydrolipoyllysine-residue acetyltransferase activity |
| Process |
| metabolic process |
| cellular metabolic process |
| organic acid metabolic process |
| oxoacid metabolic process |
| carboxylic acid metabolic process |
| monocarboxylic acid metabolic process |
| pyruvate metabolic process |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Citric Acid Cycle | 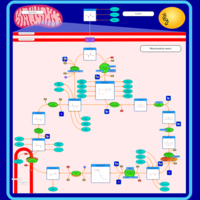 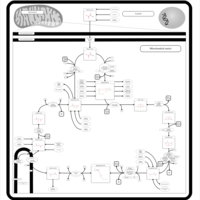 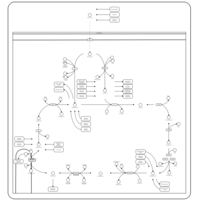 |  | | Glycolysis / Gluconeogenesis | Not Available |  | | Citrate cycle (TCA cycle) | Not Available |  | | Congenital lactic acidosis | 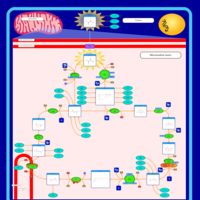 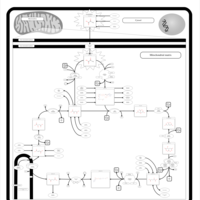 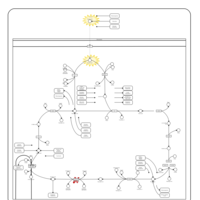 | Not Available | | Fumarase deficiency | 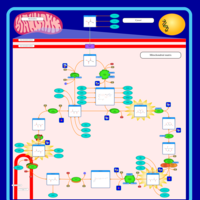 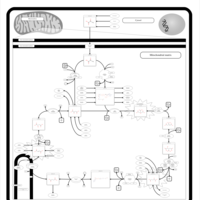  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 11 |
| Locus
| 11q23.1 |
| SNPs
| DLAT |
| Gene Sequence
|
>1944 bp
ATGTGGCGCGTCTGTGCGCGACGGGCTCAGAATGTAGCCCCATGGGCGGGACTCGAGGCT
CGGTGGACGGCCTTGCAGGAGGTACCCGGAACTCCACGAGTGACCTCGCGATCTGGCCCG
GCTCCCGTTCGTCGCAACAGCGTGACTACAGGGTATGGCGGGGTCCGGGCACTGTGCGGC
TGGACCCCCAGTTCTGGGGCCACGCCGCGGAACCGCTTACTGCTGCAGCTTTTGGGGTCG
CCCGGCCGCCGCTATTACAGTCTTCCCCCGCATCAGAAGGTTCCATTGCCTTCTCTTTCC
CCCACAATGCAGGCAGGCACCATAGCCCGTTGGGAAAAAAAAGAGGGAGACAAAATCAAT
GAAGGTGACCTAATTGCAGAGGTTGAAACTGATAAAGCCACTGTTGGATTTGAGAGCCTG
GAGGAGTGTTATATGGCAAAGATACTTGTTGCTGAAGGTACCAGGGATGTTCCCATCGGA
GCGATCATCTGTATCACAGTTGGCAAGCCTGAGGATATTGAGGCCTTTAAAAATTATACA
CTGGATTCCTCAGCAGCACCTACCCCACAAGCGGCCCCAGCACCAACCCCTGCTGCCACT
GCTTCGCCACCTACACCTTCTGCTCAGGCTCCTGGTAGCTCATATCCCCCTCACATGCAG
GTACTTCTTCCTGCCCTCTCTCCCACCATGACCATGGGCACAGTTCAGAGATGGGAAAAA
AAAGTGGGTGAGAAGCTAAGTGAAGGAGACTTACTGGCAGAGATAGAAACTGACAAAGCC
ACTATAGGTTTTGAAGTACAGGAAGAAGGTTATCTGGCAAAAATCCTGGTCCCTGAAGGC
ACAAGAGATGTCCCTCTAGGAACCCCACTCTGTATCATTGTAGAAAAAGAGGCAGATATA
TCAGCATTTGCTGACTATAGGCCAACCGAAGTAACAGATTTAAAACCACAAGCGCCACCA
CCTACCCCACCCCCGGTGGCCGCTGTTCCTCCAACTCCCCAGCCTTTAGCTCCTACACCT
TCAGCACCCTGCCCAGCTACTCCTGCTGGACCAAAGGGAAGGGTGTTTGTTAGCCCTCTT
GCAAAGAAGTTGGCAGTAGAGAAAGGGATTGATCTTACACAAGTAAAAGGGACAGGACCA
GATGGTAGAATCACCAAGAAGGATATCGACTCTTTTGTGCCTAGTAAAGTTGCTCCTGCT
CCGGCAGCTGTTGTGCCTCCCACAGGTCCTGGAATGGCACCAGTTCCTACAGGTGTCTTC
ACAGATATCCCAATCAGCAACATTCGTCGGGTTATTGCACAGCGATTAATGCAATCAAAG
CAAACCATACCTCATTATTACCTTTCTATCAATGTAAATATGGGAGAAGTTTTGTTGGTA
CGGAAAGAACTTAATAAGATATTAGAAGGGAGAAGCAAAATTTCTGTCAATGACTTCATC
ATAAAAGCTTCAGCTTTGGCATGTTTAAAAGTTCCCGAAGCAAATTCTTCTTGGATGGAC
ACAGTTATAAGACAAAATCATGTTGTTGATGTCAGTGTTGCGGTCAGTACTCCTGCAGGA
CTCATCACACCTATTGTGTTTAATGCACATATAAAAGGAGTGGAAACCATTGCTAATGAT
GTTGTTTCTTTAGCAACCAAAGCAAGAGAGGGTAAACTACAGCCACATGAATTCCAGGGT
GGCACTTTTACGATCTCCAATTTAGGAATGTTTGGAATTAAGAATTTCTCTGCTATTATT
AACCCACCTCAAGCATGTATTTTGGCAATTGGTGCTTCAGAGGATAAACTGGTCCCTGCA
GATAATGAAAAAGGGTTTGATGTGGCTAGCATGATGTCTGTTACACTCAGTTGTGATCAC
CGGGTGGTGGATGGAGCAGTTGGAGCCCAGTGGCTTGCTGAGTTTAGAAAGTACCTTGAA
AAACCTATCACTATGTTGTTGTAA
|
| Protein Properties |
|---|
| Number of Residues
| 647 |
| Molecular Weight
| 68996.03 |
| Theoretical pI
| 7.835 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial
MWRVCARRAQNVAPWAGLEARWTALQEVPGTPRVTSRSGPAPARRNSVTTGYGGVRALCG
WTPSSGATPRNRLLLQLLGSPGRRYYSLPPHQKVPLPSLSPTMQAGTIARWEKKEGDKIN
EGDLIAEVETDKATVGFESLEECYMAKILVAEGTRDVPIGAIICITVGKPEDIEAFKNYT
LDSSAAPTPQAAPAPTPAATASPPTPSAQAPGSSYPPHMQVLLPALSPTMTMGTVQRWEK
KVGEKLSEGDLLAEIETDKATIGFEVQEEGYLAKILVPEGTRDVPLGTPLCIIVEKEADI
SAFADYRPTEVTDLKPQVPPPTPPPVAAVPPTPQPLAPTPSAPCPATPAGPKGRVFVSPL
AKKLAVEKGIDLTQVKGTGPDGRITKKDIDSFVPSKVAPAPAAVVPPTGPGMAPVPTGVF
TDIPISNIRRVIAQRLMQSKQTIPHYYLSIDVNMGEVLLVRKELNKILEGRSKISVNDFI
IKASALACLKVPEANSSWMDTVIRQNHVVDVSVAVSTPAGLITPIVFNAHIKGVETIAND
VVSLATKAREGKLQPHEFQGGTFTISNLGMFGIKNFSAIINPPQACILAIGASEDKLVPA
DNEKGFDVASMMSVTLSCDHRVVDGAVGAQWLAEFRKYLEKPITMLL
|
| External Links |
|---|
| GenBank ID Protein
| 62898924 |
| UniProtKB/Swiss-Prot ID
| P10515 |
| UniProtKB/Swiss-Prot Entry Name
| ODP2_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AK223596 |
| GeneCard ID
| DLAT |
| GenAtlas ID
| DLAT |
| HGNC ID
| HGNC:2896 |
| References |
|---|
| General References
| Not Available |

